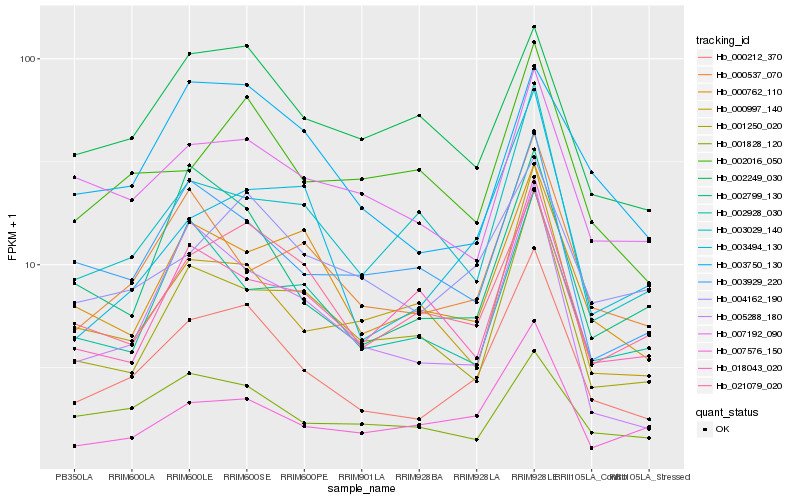
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_370 |
0.0 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 2 |
Hb_021079_020 |
0.1390958609 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 3 |
Hb_004162_190 |
0.1444223849 |
- |
- |
PREDICTED: uncharacterized protein LOC105627993 [Jatropha curcas] |
| 4 |
Hb_005288_180 |
0.1506121918 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_007576_150 |
0.1508127886 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002799_130 |
0.1516392207 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 7 |
Hb_003494_130 |
0.1517988174 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 8 |
Hb_000997_140 |
0.1552631651 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001250_020 |
0.1553838858 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003929_220 |
0.1561512645 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000537_070 |
0.1563305501 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 12 |
Hb_003750_130 |
0.1566989548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000762_110 |
0.1571956121 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002016_050 |
0.1572521549 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007192_090 |
0.1586838021 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 16 |
Hb_001828_120 |
0.1630962744 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 17 |
Hb_003029_140 |
0.1631342408 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002928_030 |
0.1690246647 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 19 |
Hb_018043_020 |
0.1691273271 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002249_030 |
0.1699204393 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |