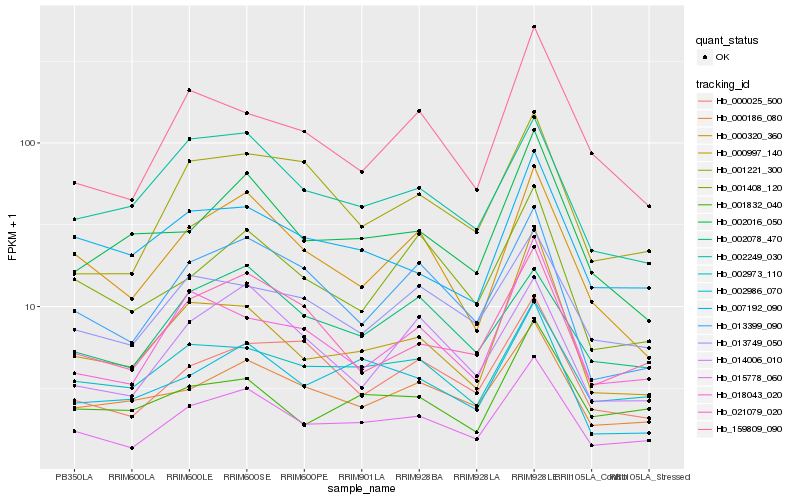
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015778_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 2 |
Hb_002973_110 |
0.0994187359 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_000186_080 |
0.1021370433 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 4 |
Hb_002986_070 |
0.1053989999 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 5 |
Hb_013399_090 |
0.1082712952 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_021079_020 |
0.1093888764 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 7 |
Hb_000320_360 |
0.1112166823 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_013749_050 |
0.1131202529 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000997_140 |
0.1155710681 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_018043_020 |
0.1193580857 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001408_120 |
0.1194423242 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_007192_090 |
0.1194686351 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 13 |
Hb_001221_300 |
0.1198442167 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002249_030 |
0.1198663741 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000025_500 |
0.1200551928 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 16 |
Hb_002016_050 |
0.120375262 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001832_040 |
0.1212244467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_014006_010 |
0.1262705326 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 19 |
Hb_002078_470 |
0.1263877524 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_159809_090 |
0.1273632107 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |