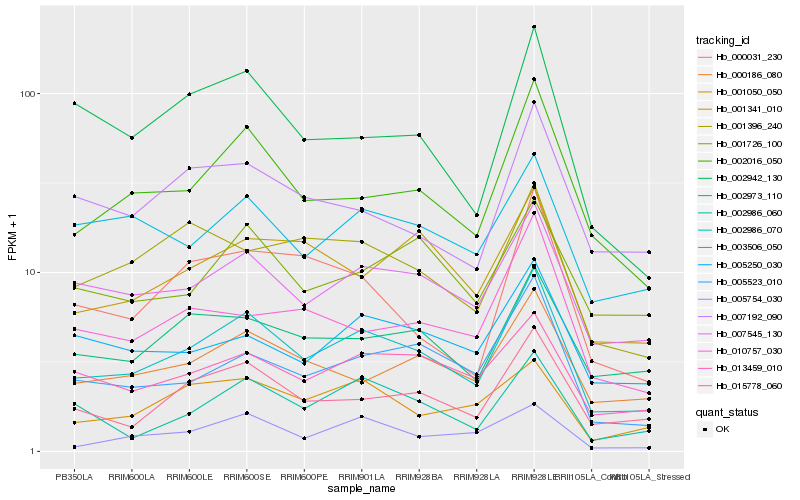
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002986_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 2 |
Hb_002016_050 |
0.0988442534 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007545_130 |
0.1026543886 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_015778_060 |
0.1053989999 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 5 |
Hb_005523_010 |
0.1174086504 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 6 |
Hb_000186_080 |
0.1190471212 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 7 |
Hb_013459_010 |
0.121328965 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 8 |
Hb_002973_110 |
0.1256867979 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_002986_060 |
0.1268196354 |
- |
- |
hypothetical protein M569_15879, partial [Genlisea aurea] |
| 10 |
Hb_001341_010 |
0.1350957615 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001050_050 |
0.1354868959 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g57250, mitochondrial [Jatropha curcas] |
| 12 |
Hb_003506_050 |
0.1372453591 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_005250_030 |
0.1379364865 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 14 |
Hb_002942_130 |
0.1379826918 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005754_030 |
0.1395587708 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 16 |
Hb_007192_090 |
0.140198499 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 17 |
Hb_001396_240 |
0.1404470224 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000031_230 |
0.1404679419 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001726_100 |
0.1408327711 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 20 |
Hb_010757_030 |
0.1412251636 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |