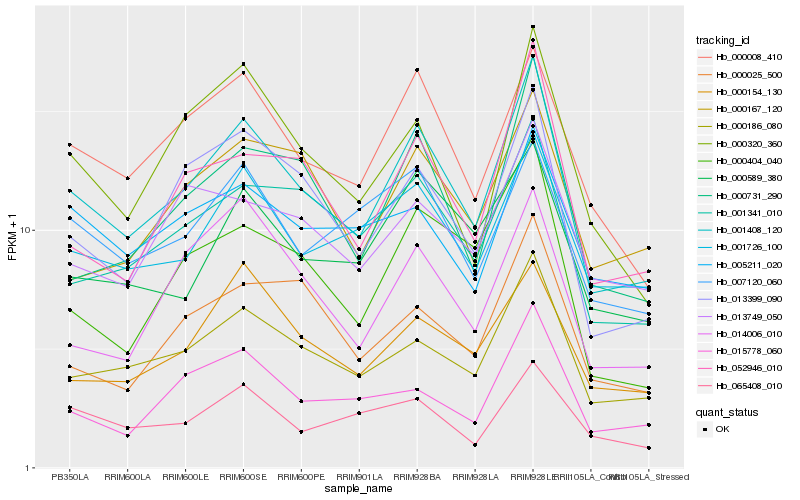
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001408_120 |
0.0 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 2 |
Hb_000186_080 |
0.0916902143 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 3 |
Hb_013399_090 |
0.0971563707 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000008_410 |
0.1020347587 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_001341_010 |
0.1077993541 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 6 |
Hb_052946_010 |
0.1091132526 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 7 |
Hb_001726_100 |
0.1094176322 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000320_360 |
0.111219442 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000731_290 |
0.1118274203 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_000404_040 |
0.1154533287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_015778_060 |
0.1194423242 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 12 |
Hb_000589_380 |
0.120460517 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 13 |
Hb_000154_130 |
0.121283049 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 14 |
Hb_007120_060 |
0.1236138782 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 15 |
Hb_000025_500 |
0.123905162 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 16 |
Hb_014006_010 |
0.124999874 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 17 |
Hb_013749_050 |
0.1269690014 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000167_120 |
0.1277191542 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 19 |
Hb_065408_010 |
0.1277821614 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 20 |
Hb_005211_020 |
0.129029397 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |