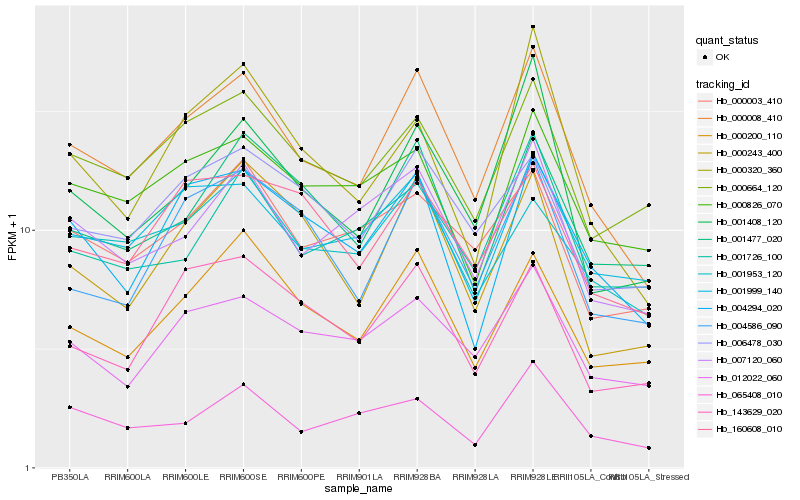
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_410 |
0.0 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_007120_060 |
0.0963098556 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 3 |
Hb_000200_110 |
0.101746421 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001408_120 |
0.1020347587 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_012022_060 |
0.10360678 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004294_020 |
0.1039379518 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001999_140 |
0.104242768 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000664_120 |
0.1047767902 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004586_090 |
0.1069764526 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006478_030 |
0.1081714476 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 11 |
Hb_000243_400 |
0.1094960766 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 12 |
Hb_001477_020 |
0.1099377267 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 13 |
Hb_065408_010 |
0.1111622065 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 14 |
Hb_160608_010 |
0.1118772181 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000320_360 |
0.112675489 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000003_410 |
0.1133791026 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 17 |
Hb_001953_120 |
0.1135743636 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 18 |
Hb_143629_020 |
0.1158072809 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 19 |
Hb_000826_070 |
0.1162337888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001726_100 |
0.1165971758 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |