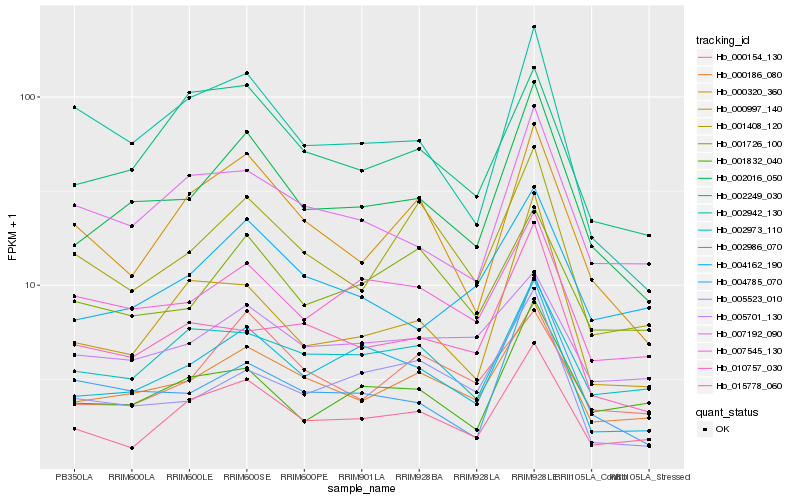
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002016_050 |
0.0 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000186_080 |
0.094516578 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 3 |
Hb_002986_070 |
0.0988442534 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 4 |
Hb_015778_060 |
0.120375262 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 5 |
Hb_007545_130 |
0.1318761653 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004785_070 |
0.1321761709 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000997_140 |
0.1335794957 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000320_360 |
0.1350804141 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_004162_190 |
0.1352821086 |
- |
- |
PREDICTED: uncharacterized protein LOC105627993 [Jatropha curcas] |
| 10 |
Hb_007192_090 |
0.1355637495 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 11 |
Hb_001832_040 |
0.1380595773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001408_120 |
0.1387068806 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_002942_130 |
0.1430383982 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 14 |
Hb_010757_030 |
0.1430503489 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_002973_110 |
0.1434483096 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_001726_100 |
0.1436537007 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002249_030 |
0.1449514824 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000154_130 |
0.1455382656 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 19 |
Hb_005523_010 |
0.1477947969 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 20 |
Hb_005701_130 |
0.1483693945 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |