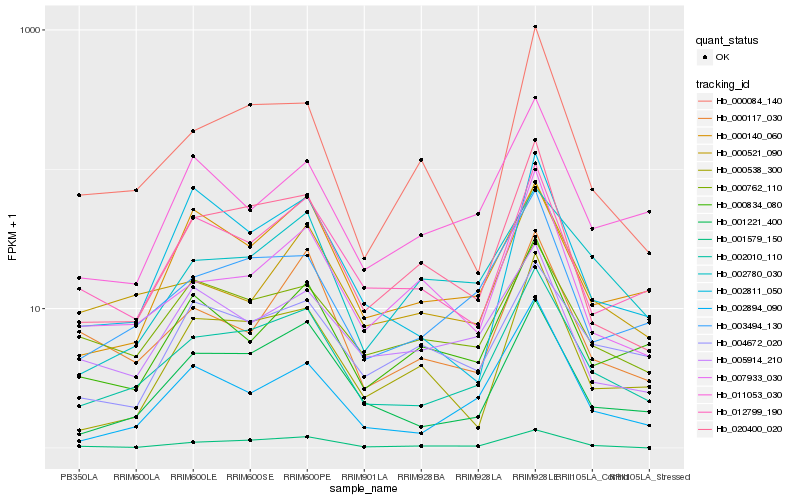
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002010_110 |
0.0 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 2 |
Hb_001221_400 |
0.1057458658 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 3 |
Hb_003494_130 |
0.1114218117 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 4 |
Hb_020400_020 |
0.1305936998 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 5 |
Hb_012799_190 |
0.1381905514 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 6 |
Hb_001579_150 |
0.1458688755 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000762_110 |
0.1459614123 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000521_090 |
0.1465666164 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002894_090 |
0.1523950706 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase [Jatropha curcas] |
| 10 |
Hb_000834_080 |
0.1531466088 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 11 |
Hb_000140_060 |
0.1543390256 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 12 |
Hb_002780_030 |
0.1570893074 |
- |
- |
PREDICTED: molybdate transporter 1 [Jatropha curcas] |
| 13 |
Hb_005914_210 |
0.1574702388 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 14 |
Hb_011053_030 |
0.1575115903 |
- |
- |
- |
| 15 |
Hb_000117_030 |
0.1585733196 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 16 |
Hb_000538_300 |
0.1593636865 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007933_030 |
0.1596145265 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 18 |
Hb_004672_020 |
0.1603335486 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 19 |
Hb_000084_140 |
0.1606159441 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 20 |
Hb_002811_050 |
0.1642183166 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |