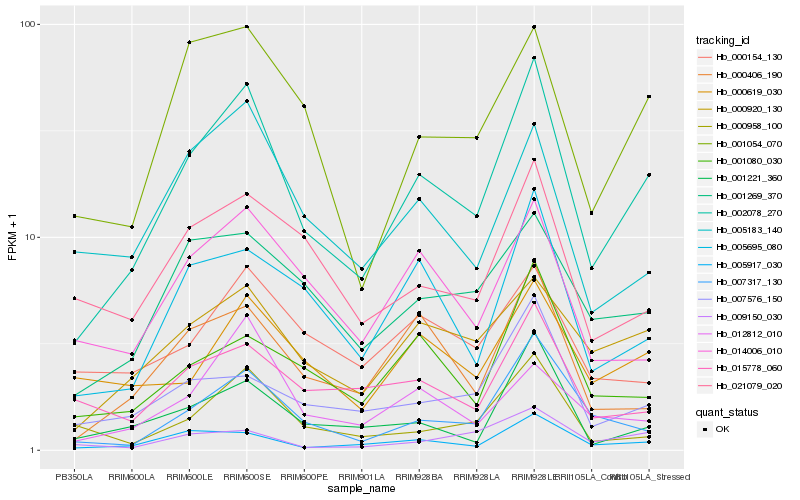
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_270 |
0.0 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 2 |
Hb_005917_030 |
0.1524008734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000920_130 |
0.1533836914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005695_080 |
0.159573064 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 5 |
Hb_007317_130 |
0.1615557265 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 6 |
Hb_001054_070 |
0.1620298984 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 7 |
Hb_000406_190 |
0.1634439972 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_001269_370 |
0.1670417082 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 9 |
Hb_001080_030 |
0.1770435875 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 10 |
Hb_007576_150 |
0.1784938514 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 11 |
Hb_014006_010 |
0.1794538316 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 12 |
Hb_001221_360 |
0.1801170228 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 13 |
Hb_000619_030 |
0.1802065729 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_021079_020 |
0.1817248919 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 15 |
Hb_005183_140 |
0.1844521458 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 16 |
Hb_015778_060 |
0.1863833667 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 17 |
Hb_000958_100 |
0.1907844545 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 18 |
Hb_012812_010 |
0.1915715448 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 19 |
Hb_000154_130 |
0.1939966157 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 20 |
Hb_009150_030 |
0.197066157 |
- |
- |
hypothetical protein POPTR_0010s14920g [Populus trichocarpa] |