| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
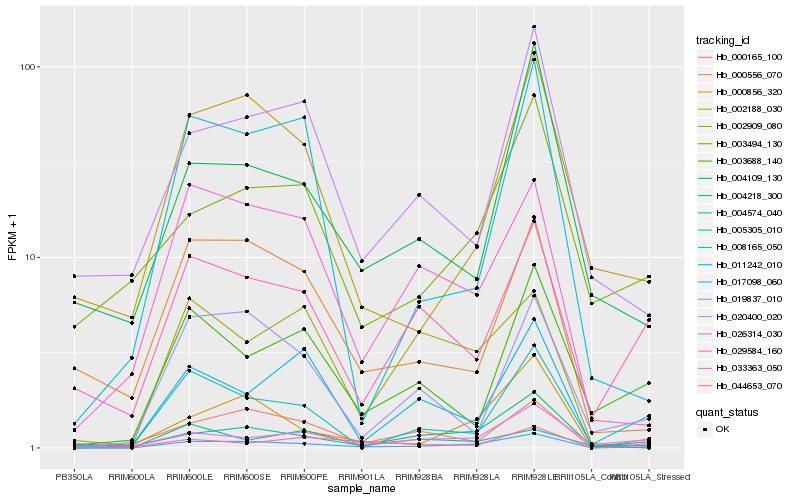
Hb_017098_060 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Pyrus x bretschneideri] |
| 2 |
Hb_000856_320 |
0.1966898284 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 3 |
Hb_000165_100 |
0.2099082294 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 4 |
Hb_029584_160 |
0.2135251339 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 5 |
Hb_026314_030 |
0.2175390376 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 6 |
Hb_005305_010 |
0.2248073295 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 7 |
Hb_004218_300 |
0.2249490943 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 8 |
Hb_002188_030 |
0.2255576846 |
- |
- |
PREDICTED: probable zinc metallopeptidase EGY3, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_033363_050 |
0.2290361511 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 isoform X2 [Jatropha curcas] |
| 10 |
Hb_011242_010 |
0.2299805058 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 11 |
Hb_004574_040 |
0.2306261941 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 12 |
Hb_019837_010 |
0.2332701276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_020400_020 |
0.2353261722 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 14 |
Hb_003494_130 |
0.2375325329 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 15 |
Hb_000556_070 |
0.2379869328 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 16 |
Hb_003688_140 |
0.2389890623 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 17 |
Hb_002909_080 |
0.2395124011 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 18 |
Hb_004109_130 |
0.2395444184 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_008165_050 |
0.2396497927 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_044653_070 |
0.2420625713 |
- |
- |
hypothetical protein CICLE_v10018540mg [Citrus clementina] |