| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
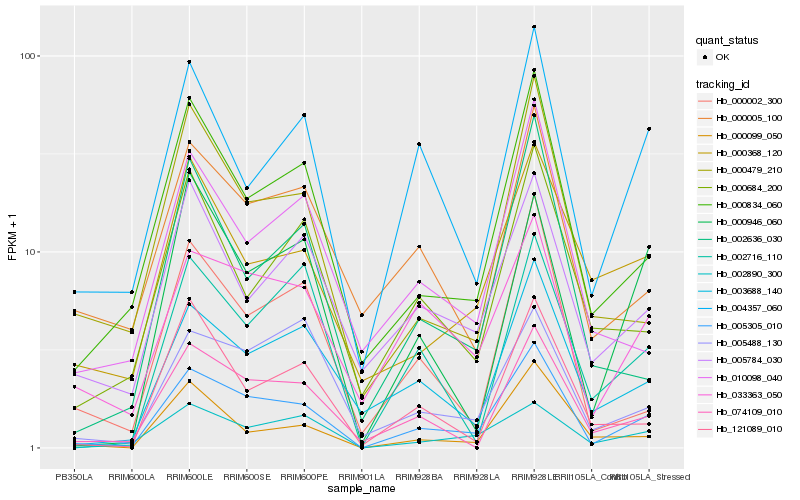
Hb_005305_010 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 2 |
Hb_000834_060 |
0.1494226949 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_033363_050 |
0.1581781306 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 isoform X2 [Jatropha curcas] |
| 4 |
Hb_074109_010 |
0.1608611229 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 5 |
Hb_003688_140 |
0.1646955628 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 6 |
Hb_004357_060 |
0.1701245807 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 7 |
Hb_000002_300 |
0.1798526688 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 8 |
Hb_000479_210 |
0.1812813473 |
- |
- |
PREDICTED: uncharacterized protein LOC105644126 [Jatropha curcas] |
| 9 |
Hb_000005_100 |
0.1823633158 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002890_300 |
0.1829666272 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 11 |
Hb_005784_030 |
0.1831571603 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000099_050 |
0.185000074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005488_130 |
0.1898183573 |
- |
- |
transporter, putative [Ricinus communis] |
| 14 |
Hb_010098_040 |
0.1909734806 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 15 |
Hb_121089_010 |
0.1918382982 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 16 |
Hb_000368_120 |
0.1923817574 |
- |
- |
PREDICTED: uncharacterized protein LOC105633862 [Jatropha curcas] |
| 17 |
Hb_002636_030 |
0.1934446159 |
- |
- |
hypothetical protein JCGZ_11800 [Jatropha curcas] |
| 18 |
Hb_000946_060 |
0.193997899 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000684_200 |
0.1942745426 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 20 |
Hb_002716_110 |
0.1957530587 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |