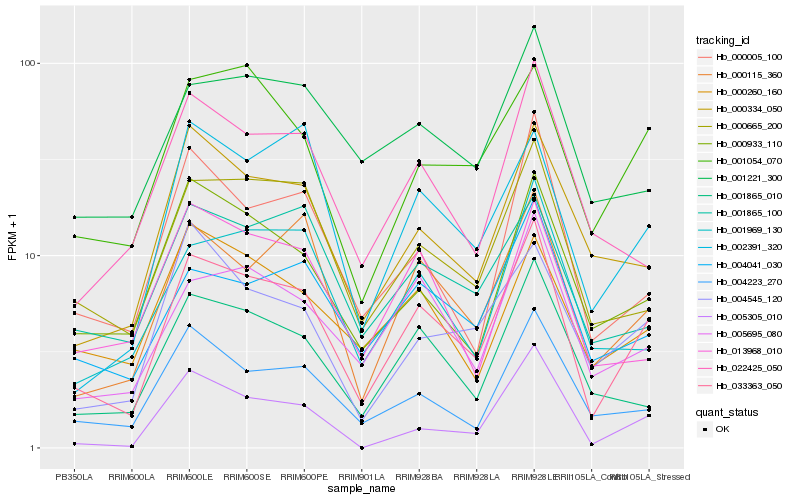
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033363_050 |
0.0 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000115_360 |
0.1324560501 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000665_200 |
0.1351377217 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_005695_080 |
0.1381295455 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 5 |
Hb_001865_010 |
0.1409793651 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 6 |
Hb_004223_270 |
0.1419221716 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000005_100 |
0.1445935679 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004041_030 |
0.1449151026 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000933_110 |
0.148101871 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 10 |
Hb_000334_050 |
0.1489415643 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 11 |
Hb_001054_070 |
0.1496947859 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 12 |
Hb_001969_130 |
0.1552877454 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 13 |
Hb_022425_050 |
0.1567790385 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 14 |
Hb_005305_010 |
0.1581781306 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 15 |
Hb_002391_320 |
0.1604183137 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 16 |
Hb_001221_300 |
0.1618691827 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000260_160 |
0.1631247705 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_001865_100 |
0.1645830191 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 19 |
Hb_013968_010 |
0.1649747894 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 20 |
Hb_004545_120 |
0.1656441042 |
- |
- |
PREDICTED: uncharacterized protein LOC105647604 [Jatropha curcas] |