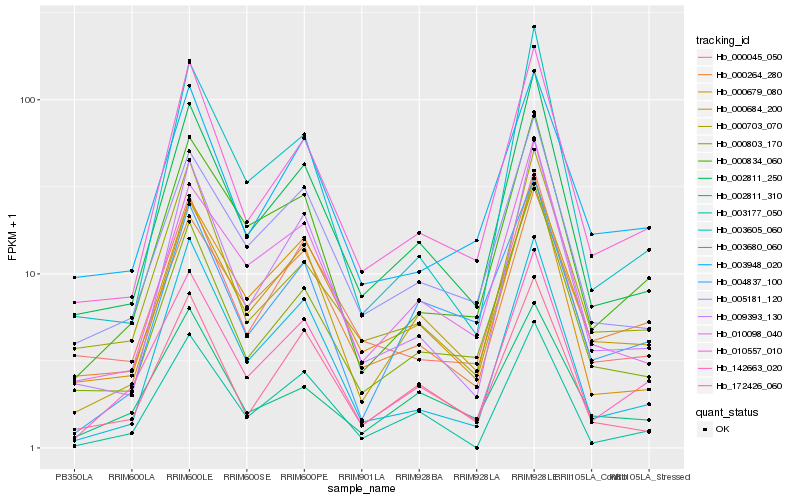
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142663_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 2 |
Hb_000834_060 |
0.0919487516 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000684_200 |
0.0985990063 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 4 |
Hb_002811_250 |
0.1002348992 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_010557_010 |
0.1042700304 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005181_120 |
0.1179243971 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_003177_050 |
0.1186506182 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Populus euphratica] |
| 8 |
Hb_000264_280 |
0.1207605564 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 9 |
Hb_009393_130 |
0.1263331588 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 10 |
Hb_000679_080 |
0.1273053304 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000045_050 |
0.1294145754 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_172426_060 |
0.1309586144 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 13 |
Hb_003948_020 |
0.1318262855 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 14 |
Hb_003605_060 |
0.1324609696 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000703_070 |
0.1331158213 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010098_040 |
0.1332117434 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 17 |
Hb_003680_060 |
0.133510844 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 18 |
Hb_002811_310 |
0.1346201165 |
- |
- |
hypothetical protein VITISV_025505 [Vitis vinifera] |
| 19 |
Hb_000803_170 |
0.1348584416 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 20 |
Hb_004837_100 |
0.1354530022 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |