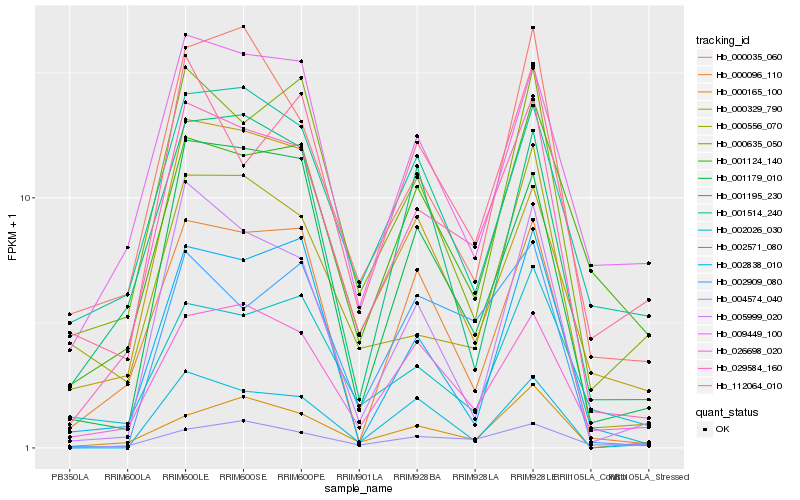
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029584_160 |
0.0 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 2 |
Hb_026698_020 |
0.1365389965 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 3 |
Hb_001179_010 |
0.1380085417 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 4 |
Hb_000035_060 |
0.1390335064 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000635_050 |
0.1402416288 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 6 |
Hb_000096_110 |
0.1422188349 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 7 |
Hb_005999_020 |
0.1513044914 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 8 |
Hb_000556_070 |
0.1533355845 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 9 |
Hb_002909_080 |
0.1533995125 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 10 |
Hb_009449_100 |
0.1574493504 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 11 |
Hb_000165_100 |
0.1589757392 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 12 |
Hb_004574_040 |
0.1610314269 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 13 |
Hb_112064_010 |
0.1615605599 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 14 |
Hb_001195_230 |
0.1633883764 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 15 |
Hb_001514_240 |
0.1644356021 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 16 |
Hb_002571_080 |
0.165349062 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |
| 17 |
Hb_002026_030 |
0.165373405 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 18 |
Hb_002838_010 |
0.1655344118 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 19 |
Hb_000329_790 |
0.1669858418 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 20 |
Hb_001124_140 |
0.1679683858 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |