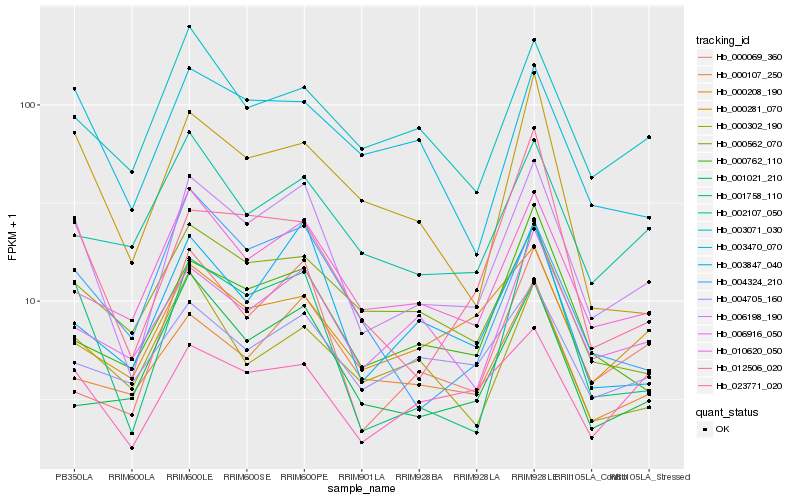
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006198_190 |
0.0 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 2 |
Hb_001758_110 |
0.1180429926 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 3 |
Hb_010620_050 |
0.1296511164 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 4 |
Hb_003847_040 |
0.1364440482 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 5 |
Hb_006916_050 |
0.1445997654 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 6 |
Hb_012506_020 |
0.1456097735 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g44820 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000069_360 |
0.1494062479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000107_250 |
0.1495483097 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 9 |
Hb_003071_030 |
0.1497413787 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000562_070 |
0.1515732712 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004324_210 |
0.1535316697 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 12 |
Hb_002107_050 |
0.1542409478 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 13 |
Hb_000762_110 |
0.1547649473 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003470_070 |
0.1553832357 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 15 |
Hb_004705_160 |
0.1593253694 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 16 |
Hb_001021_210 |
0.1601596677 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_023771_020 |
0.1612119251 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000208_190 |
0.1615437346 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 19 |
Hb_000302_190 |
0.1620190938 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 20 |
Hb_000281_070 |
0.1620277345 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |