| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
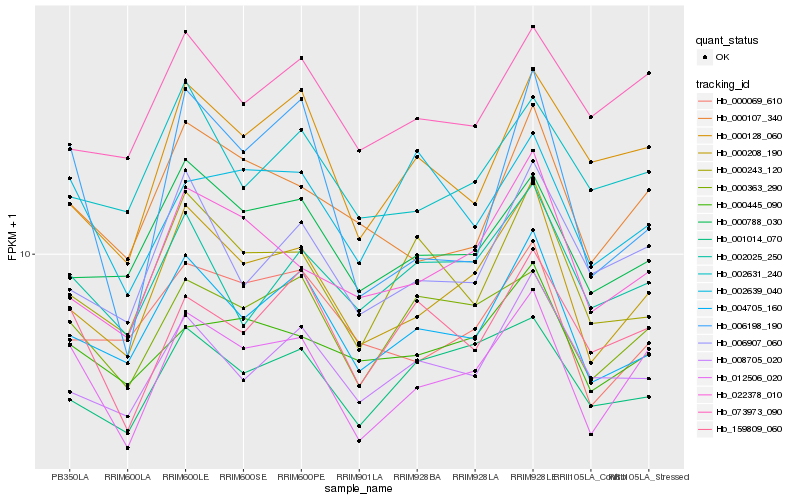
Hb_012506_020 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g44820 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000208_190 |
0.0963591012 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 3 |
Hb_000363_290 |
0.119142824 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 4 |
Hb_004705_160 |
0.1238409573 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 5 |
Hb_000107_340 |
0.1241621189 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 6 |
Hb_159809_060 |
0.124641034 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_022378_010 |
0.1354909374 |
- |
- |
PREDICTED: uncharacterized protein LOC105634715 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006907_060 |
0.1392005303 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000445_090 |
0.139246908 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 10 |
Hb_000788_030 |
0.1394415659 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 11 |
Hb_000128_060 |
0.1396520605 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 12 |
Hb_001014_070 |
0.1413047083 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 13 |
Hb_073973_090 |
0.1444591362 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 14 |
Hb_002025_250 |
0.144677173 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 15 |
Hb_000069_610 |
0.144729418 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 16 |
Hb_000243_120 |
0.1452759429 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002639_040 |
0.1454645534 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 18 |
Hb_006198_190 |
0.1456097735 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 19 |
Hb_008705_020 |
0.1479380682 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_002631_240 |
0.1482338802 |
- |
- |
JHL17M24.3 [Jatropha curcas] |