| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
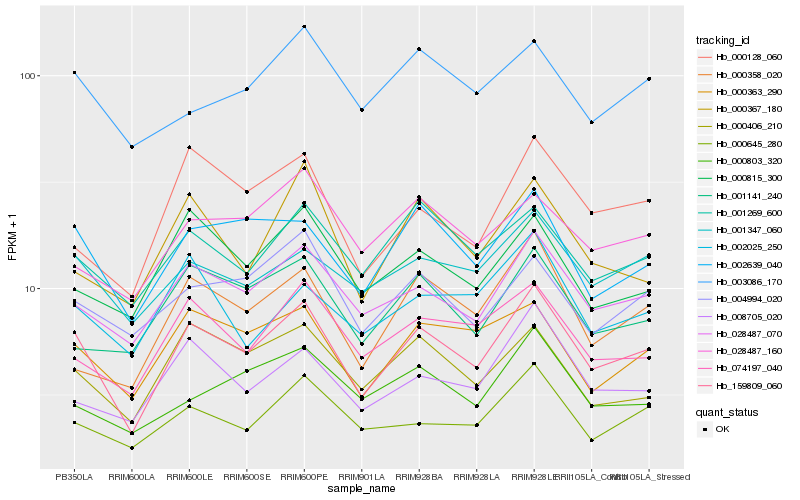
Hb_159809_060 |
0.0 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_028487_070 |
0.086406529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002639_040 |
0.0966499273 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 4 |
Hb_000363_290 |
0.1007954689 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 5 |
Hb_001347_060 |
0.1036929616 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_000645_280 |
0.1049808338 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 7 |
Hb_001269_600 |
0.1067033842 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 8 |
Hb_000406_210 |
0.106711153 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 9 |
Hb_074197_040 |
0.1074139778 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 10 |
Hb_008705_020 |
0.1113249924 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_000128_060 |
0.1124733876 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 12 |
Hb_004994_020 |
0.1126272996 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 13 |
Hb_000815_300 |
0.1159928427 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 14 |
Hb_001141_240 |
0.1175644578 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000358_020 |
0.117603256 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 16 |
Hb_003086_170 |
0.1192065574 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 17 |
Hb_002025_250 |
0.1199038382 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 18 |
Hb_000803_320 |
0.1210255913 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 19 |
Hb_028487_160 |
0.1214306407 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 20 |
Hb_000367_180 |
0.1225192606 |
- |
- |
Heparanase-2, putative [Ricinus communis] |