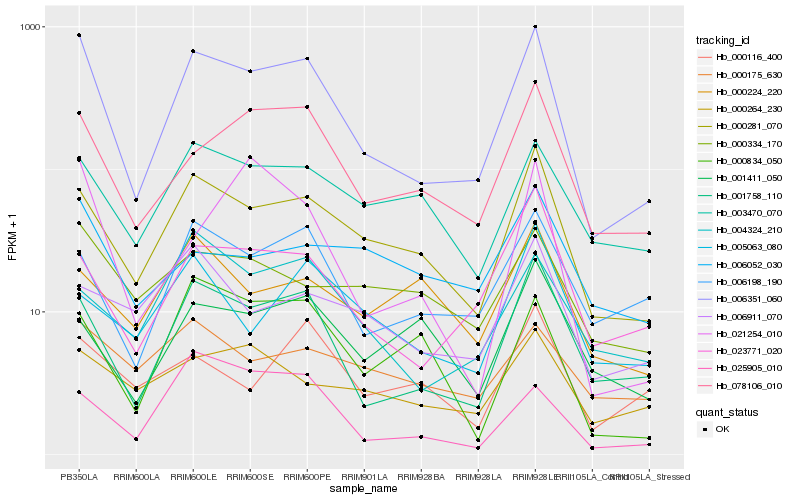
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006351_060 |
0.0 |
- |
- |
PREDICTED: 17.8 kDa class I heat shock protein-like [Cucumis sativus] |
| 2 |
Hb_001758_110 |
0.1314938401 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 3 |
Hb_000281_070 |
0.1374098576 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 4 |
Hb_078106_010 |
0.1582468213 |
- |
- |
seed maturation protein PM37 [Populus trichocarpa] |
| 5 |
Hb_023771_020 |
0.1756180141 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X3 [Jatropha curcas] |
| 6 |
Hb_006198_190 |
0.1761163415 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 7 |
Hb_003470_070 |
0.1957351645 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 8 |
Hb_004324_210 |
0.2046655878 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 9 |
Hb_000834_050 |
0.2092128637 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 10 |
Hb_000334_170 |
0.2097279567 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000264_230 |
0.2099628737 |
- |
- |
PREDICTED: uncharacterized protein LOC105649528 [Jatropha curcas] |
| 12 |
Hb_021254_010 |
0.2117691957 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 13 |
Hb_025905_010 |
0.2143009828 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 14 |
Hb_000175_630 |
0.2145196789 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g11310, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000116_400 |
0.2178435041 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_006052_030 |
0.2182979628 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A [Jatropha curcas] |
| 17 |
Hb_001411_050 |
0.2185551873 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 18 |
Hb_006911_070 |
0.2187982401 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 19 |
Hb_005063_080 |
0.2208885663 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000224_220 |
0.2251029395 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |