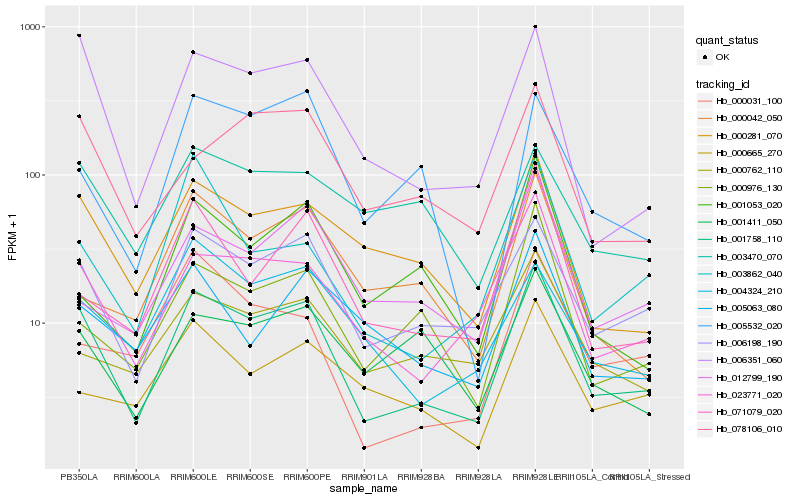
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001758_110 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_006198_190 |
0.1180429926 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 3 |
Hb_006351_060 |
0.1314938401 |
- |
- |
PREDICTED: 17.8 kDa class I heat shock protein-like [Cucumis sativus] |
| 4 |
Hb_023771_020 |
0.1399873114 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000281_070 |
0.147110742 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 6 |
Hb_078106_010 |
0.170160806 |
- |
- |
seed maturation protein PM37 [Populus trichocarpa] |
| 7 |
Hb_000976_130 |
0.1784843606 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 8 |
Hb_001411_050 |
0.1821315629 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 9 |
Hb_004324_210 |
0.1845757858 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 10 |
Hb_071079_020 |
0.1854547293 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 11 |
Hb_000762_110 |
0.1865191377 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_012799_190 |
0.1870366254 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 13 |
Hb_005063_080 |
0.1881264502 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005532_020 |
0.1887400427 |
- |
- |
PREDICTED: uncharacterized protein LOC100262861 [Vitis vinifera] |
| 15 |
Hb_000665_270 |
0.1899615344 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 16 |
Hb_003862_040 |
0.1943483185 |
- |
- |
Plastid-specific 30S ribosomal protein 1, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000042_050 |
0.1949623057 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001053_020 |
0.1959317967 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 19 |
Hb_000031_100 |
0.1959647529 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 20 |
Hb_003470_070 |
0.1968538874 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |