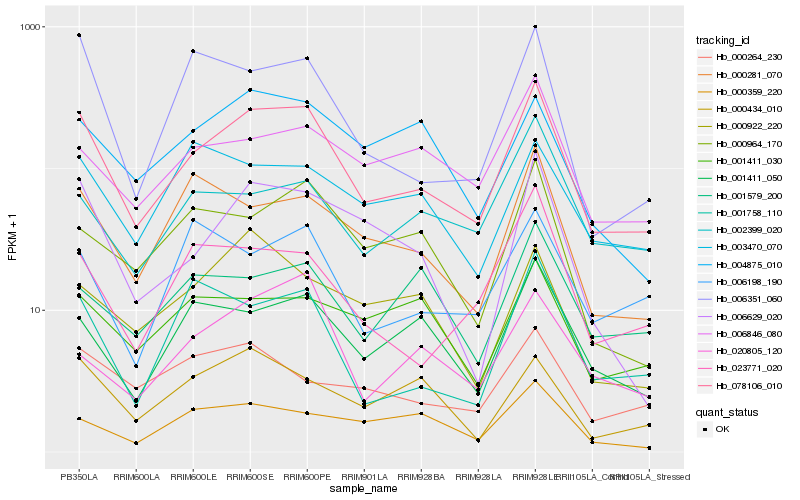
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078106_010 |
0.0 |
- |
- |
seed maturation protein PM37 [Populus trichocarpa] |
| 2 |
Hb_001411_050 |
0.1559520595 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 3 |
Hb_006351_060 |
0.1582468213 |
- |
- |
PREDICTED: 17.8 kDa class I heat shock protein-like [Cucumis sativus] |
| 4 |
Hb_006629_020 |
0.1619233449 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 5 |
Hb_000281_070 |
0.1619547317 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 6 |
Hb_006846_080 |
0.1654333843 |
- |
- |
calnexin, putative [Ricinus communis] |
| 7 |
Hb_023771_020 |
0.1672073514 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000964_170 |
0.1699658446 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 9 |
Hb_001758_110 |
0.170160806 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 10 |
Hb_002399_020 |
0.1732600375 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 11 |
Hb_004875_010 |
0.1747000001 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 12 |
Hb_000434_010 |
0.1751729879 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 13 |
Hb_000264_230 |
0.1761630536 |
- |
- |
PREDICTED: uncharacterized protein LOC105649528 [Jatropha curcas] |
| 14 |
Hb_006198_190 |
0.1769922663 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 15 |
Hb_003470_070 |
0.179323681 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 16 |
Hb_001411_030 |
0.1793764198 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 17 |
Hb_020805_120 |
0.1805524208 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 18 |
Hb_001579_200 |
0.1823457319 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 19 |
Hb_000922_220 |
0.1826615331 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 20 |
Hb_000359_220 |
0.1832028346 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |