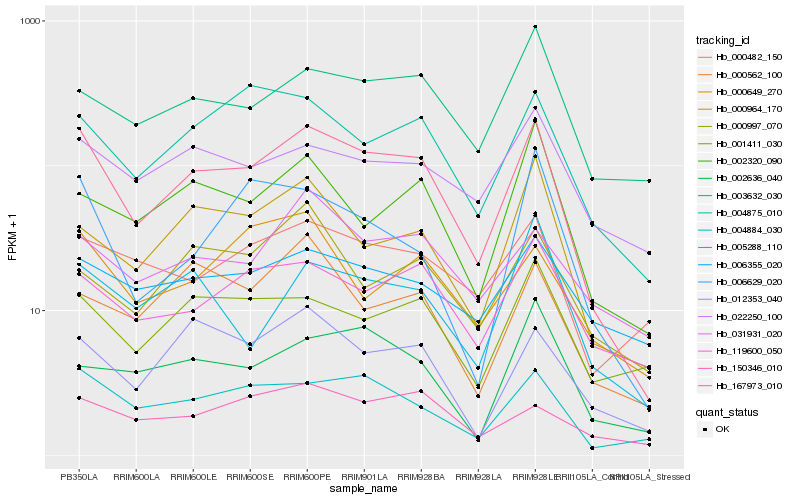
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_167973_010 |
0.0 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 2 |
Hb_004884_030 |
0.1386062789 |
- |
- |
PREDICTED: ammonium transporter 3 member 3 [Vitis vinifera] |
| 3 |
Hb_004875_010 |
0.1586923337 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 4 |
Hb_000964_170 |
0.1608006029 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 5 |
Hb_012353_040 |
0.1624654923 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_119600_050 |
0.1628687615 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_003632_030 |
0.1679753383 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 8 |
Hb_000482_150 |
0.1695524872 |
- |
- |
PREDICTED: zinc transporter 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001411_030 |
0.169665415 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 10 |
Hb_022250_100 |
0.1734468085 |
- |
- |
calnexin, putative [Ricinus communis] |
| 11 |
Hb_002320_090 |
0.174103029 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 12 |
Hb_031931_020 |
0.1768654406 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 13 |
Hb_150346_010 |
0.1782130337 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 14 |
Hb_006355_020 |
0.1789102655 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 15 |
Hb_000562_100 |
0.1790274757 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 16 |
Hb_005288_110 |
0.1805842258 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 17 |
Hb_006629_020 |
0.1809257292 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 18 |
Hb_000997_070 |
0.1839799462 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 19 |
Hb_000649_270 |
0.1844034413 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_002636_040 |
0.1845925304 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |