| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
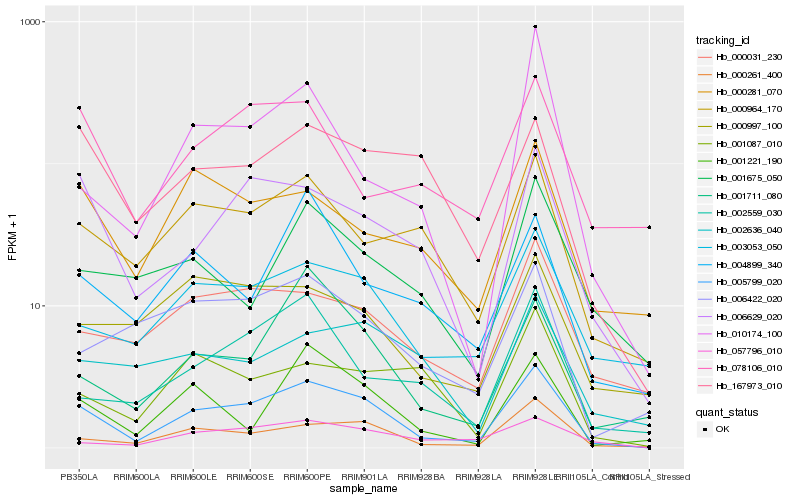
Hb_005799_020 |
0.0 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET6b [Jatropha curcas] |
| 2 |
Hb_000261_400 |
0.1526517089 |
- |
- |
hypothetical protein JCGZ_01125 [Jatropha curcas] |
| 3 |
Hb_003053_050 |
0.1818214644 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 8 [Jatropha curcas] |
| 4 |
Hb_006629_020 |
0.1824990944 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 5 |
Hb_000964_170 |
0.1951202425 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 6 |
Hb_000031_230 |
0.1953072668 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 7 |
Hb_057796_010 |
0.2049297594 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 8 |
Hb_002559_030 |
0.2086267684 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 9 |
Hb_001711_080 |
0.2091852597 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 10 |
Hb_001221_190 |
0.2098151327 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 11 |
Hb_006422_020 |
0.2132435771 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 12 |
Hb_000281_070 |
0.2137779321 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 13 |
Hb_167973_010 |
0.2140516581 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 14 |
Hb_000997_100 |
0.2185429244 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004899_340 |
0.2193557094 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 16 |
Hb_001675_050 |
0.2243846564 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 17 |
Hb_001087_010 |
0.2252606572 |
- |
- |
PREDICTED: uncharacterized protein LOC105630042 [Jatropha curcas] |
| 18 |
Hb_078106_010 |
0.2261403931 |
- |
- |
seed maturation protein PM37 [Populus trichocarpa] |
| 19 |
Hb_002636_040 |
0.2266510927 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010174_100 |
0.2287454593 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |