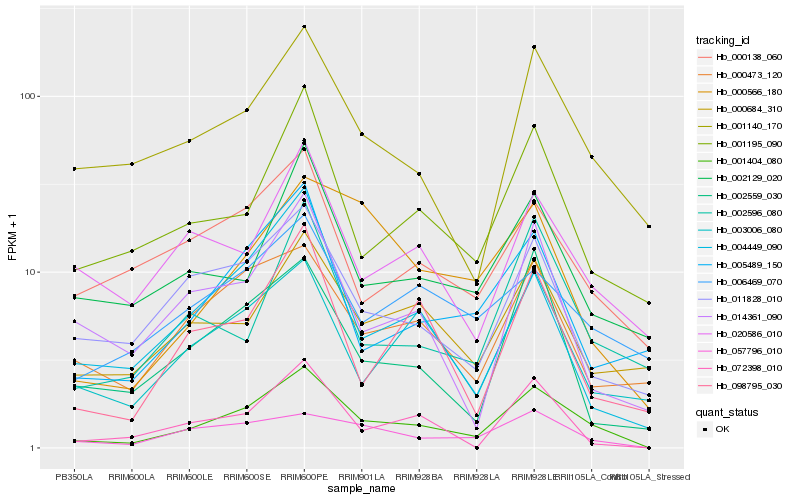
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001404_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_057796_010 |
0.1758646038 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 3 |
Hb_001140_170 |
0.1796415237 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 4 |
Hb_001195_090 |
0.185702489 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 5 |
Hb_011828_010 |
0.1869239696 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 6 |
Hb_005489_150 |
0.1933237228 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 7 |
Hb_000473_120 |
0.1936149251 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 8 |
Hb_003006_080 |
0.194068501 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000684_310 |
0.1945851997 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 10 |
Hb_002559_030 |
0.1959428799 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 11 |
Hb_004449_090 |
0.1996817241 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 12 |
Hb_002129_020 |
0.2001965529 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 13 |
Hb_098795_030 |
0.2008443221 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 14 |
Hb_002596_080 |
0.2014509927 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 15 |
Hb_000566_180 |
0.202197584 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor 7-like [Jatropha curcas] |
| 16 |
Hb_000138_060 |
0.203529674 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 17 |
Hb_014361_090 |
0.2040465826 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 18 |
Hb_020586_010 |
0.2042874169 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_006469_070 |
0.2045955066 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 20 |
Hb_072398_010 |
0.2055145836 |
- |
- |
pepsin A, putative [Ricinus communis] |