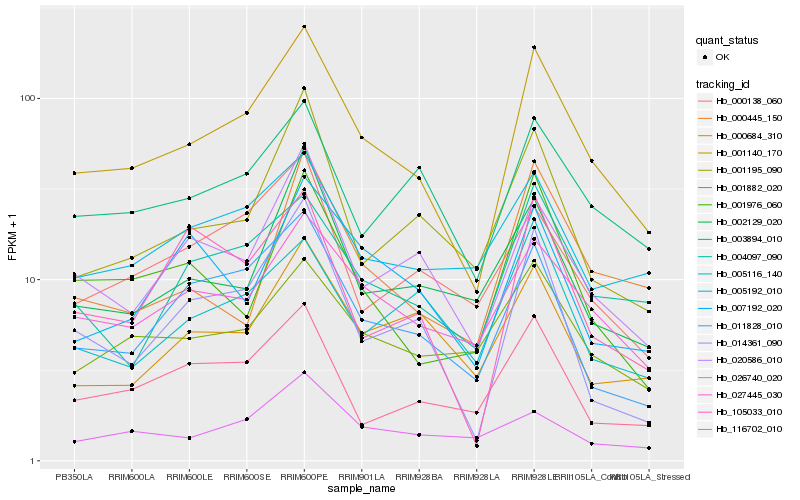
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001140_170 |
0.0 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 2 |
Hb_027445_030 |
0.1241733408 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_011828_010 |
0.1303799948 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 4 |
Hb_020586_010 |
0.1320674505 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_004097_090 |
0.1381013444 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 6 |
Hb_014361_090 |
0.1397701352 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 7 |
Hb_003894_010 |
0.1417417348 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_001882_020 |
0.1457334247 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 9 |
Hb_001195_090 |
0.1487424692 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 10 |
Hb_116702_010 |
0.1525535204 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 11 |
Hb_002129_020 |
0.153285355 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 12 |
Hb_005192_010 |
0.154307424 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_001976_060 |
0.1543785141 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000138_060 |
0.1551632756 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 15 |
Hb_000684_310 |
0.1557795149 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 16 |
Hb_000445_150 |
0.1594545549 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 17 |
Hb_005116_140 |
0.1609413075 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 18 |
Hb_105033_010 |
0.1611336455 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007192_020 |
0.1624516377 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 20 |
Hb_026740_020 |
0.163506431 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |