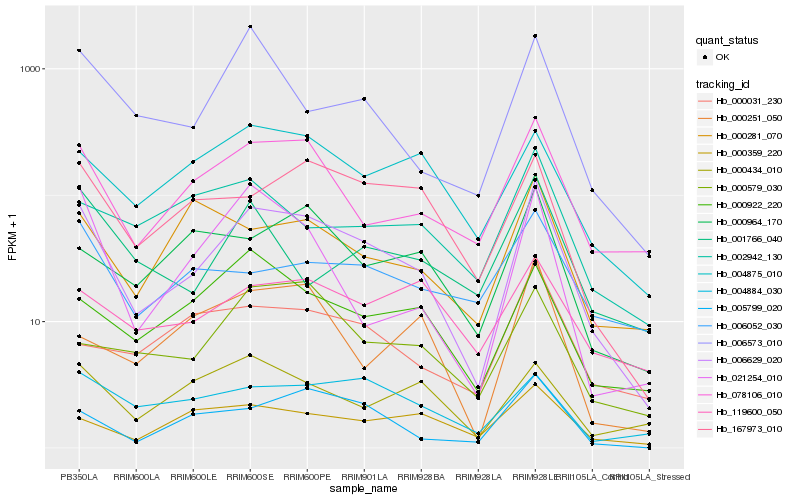
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006629_020 |
0.0 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 2 |
Hb_078106_010 |
0.1619233449 |
- |
- |
seed maturation protein PM37 [Populus trichocarpa] |
| 3 |
Hb_167973_010 |
0.1809257292 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 4 |
Hb_005799_020 |
0.1824990944 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET6b [Jatropha curcas] |
| 5 |
Hb_006573_010 |
0.1930767623 |
- |
- |
heat shock protein [Hevea brasiliensis] |
| 6 |
Hb_000922_220 |
0.1942991944 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 7 |
Hb_004875_010 |
0.1970216221 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 8 |
Hb_021254_010 |
0.1994573324 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 9 |
Hb_004884_030 |
0.2016788679 |
- |
- |
PREDICTED: ammonium transporter 3 member 3 [Vitis vinifera] |
| 10 |
Hb_000359_220 |
0.2027211945 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000964_170 |
0.2042149682 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 12 |
Hb_000579_030 |
0.2073681809 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 13 |
Hb_000434_010 |
0.2084824418 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 14 |
Hb_000281_070 |
0.2119218617 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 15 |
Hb_006052_030 |
0.2131808902 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A [Jatropha curcas] |
| 16 |
Hb_119600_050 |
0.2140200048 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_001766_040 |
0.2146583967 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 18 |
Hb_000031_230 |
0.2160992712 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002942_130 |
0.2166200638 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000251_050 |
0.2190641104 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |