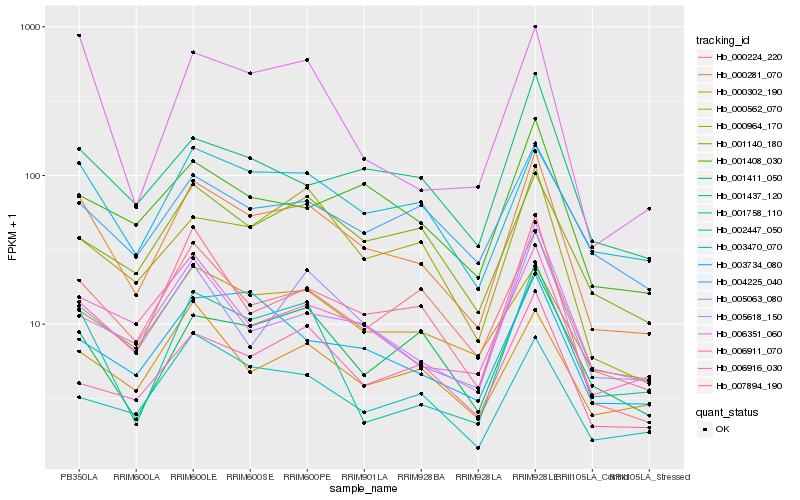
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000281_070 |
0.0 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 2 |
Hb_000224_220 |
0.1200441519 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |
| 3 |
Hb_003470_070 |
0.1225019121 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 4 |
Hb_006911_070 |
0.1307154791 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 5 |
Hb_005063_080 |
0.1341690548 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001411_050 |
0.1345846006 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 7 |
Hb_001408_030 |
0.1370300509 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006351_060 |
0.1374098576 |
- |
- |
PREDICTED: 17.8 kDa class I heat shock protein-like [Cucumis sativus] |
| 9 |
Hb_000964_170 |
0.1389278276 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 10 |
Hb_001437_120 |
0.1396938217 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 11 |
Hb_001140_180 |
0.1407220355 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 12 |
Hb_007894_190 |
0.141077701 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 13 |
Hb_000562_070 |
0.1417663505 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000302_190 |
0.1420072683 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 15 |
Hb_004225_040 |
0.1429886372 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 16 |
Hb_003734_080 |
0.1449829106 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 17 |
Hb_001758_110 |
0.147110742 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 18 |
Hb_006916_030 |
0.1471693462 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 19 |
Hb_002447_050 |
0.1471696509 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_005618_150 |
0.1480345591 |
- |
- |
PREDICTED: protein translocase subunit SecA, chloroplastic [Jatropha curcas] |