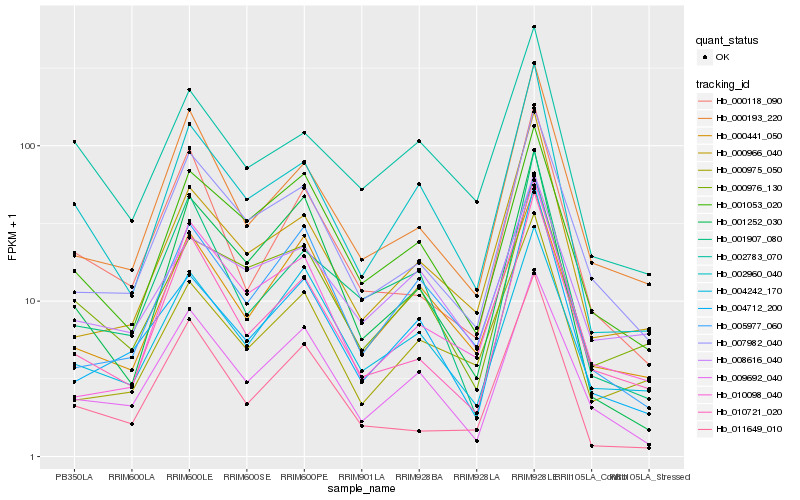
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002960_040 |
0.0 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 2 |
Hb_002783_070 |
0.0965512625 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 3 |
Hb_000976_130 |
0.1202800979 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 4 |
Hb_000966_040 |
0.1238893433 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 5 |
Hb_001907_080 |
0.1250429435 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 6 |
Hb_009692_040 |
0.1252165875 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000193_220 |
0.1294468879 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001053_020 |
0.1331900682 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 9 |
Hb_001252_030 |
0.1346785982 |
- |
- |
Serine/threonine-protein kinase Nek8, putative [Ricinus communis] |
| 10 |
Hb_010721_020 |
0.1364853756 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007982_040 |
0.1377874198 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004712_200 |
0.1383909995 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 13 |
Hb_000441_050 |
0.1399096036 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 14 |
Hb_005977_060 |
0.1426588666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000975_050 |
0.1440741837 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000118_090 |
0.1502191649 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_004242_170 |
0.1503000111 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 18 |
Hb_011649_010 |
0.1512203847 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 19 |
Hb_008616_040 |
0.1518340812 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 20 |
Hb_010098_040 |
0.1536103162 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |