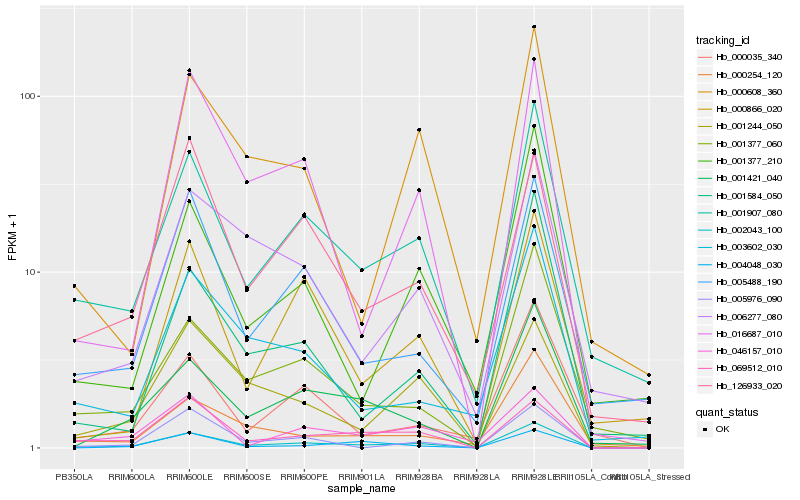
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002043_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001907_080 |
0.1221089782 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 3 |
Hb_001421_040 |
0.1479536798 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 2 [Jatropha curcas] |
| 4 |
Hb_069512_010 |
0.1535389374 |
- |
- |
- |
| 5 |
Hb_001377_060 |
0.1651096552 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 6 |
Hb_016687_010 |
0.1657912707 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 7 |
Hb_005488_190 |
0.1706871236 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 8 |
Hb_001244_050 |
0.1750684861 |
- |
- |
PREDICTED: uncharacterized protein LOC105643334 [Jatropha curcas] |
| 9 |
Hb_126933_020 |
0.1797558205 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 10 |
Hb_046157_010 |
0.1819869851 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 11 |
Hb_000866_020 |
0.1824750744 |
- |
- |
hypothetical protein B456_004G042000 [Gossypium raimondii] |
| 12 |
Hb_005976_090 |
0.1828976635 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001584_050 |
0.1830365478 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001377_210 |
0.1836079838 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 15 |
Hb_000254_120 |
0.1842994617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000035_340 |
0.1843248973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004048_030 |
0.1847700522 |
- |
- |
hypothetical protein B456_009G371200, partial [Gossypium raimondii] |
| 18 |
Hb_003602_030 |
0.1859330133 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 19 |
Hb_000608_360 |
0.1866137887 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 20 |
Hb_006277_080 |
0.1870388735 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |