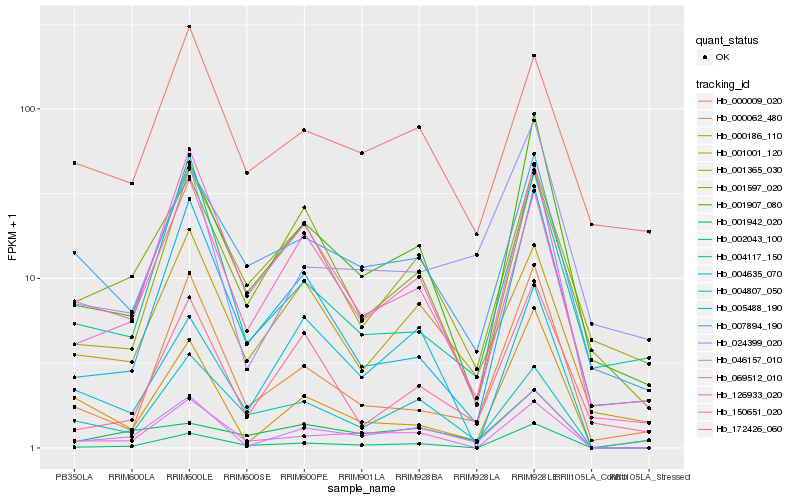
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046157_010 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 2 |
Hb_069512_010 |
0.1610694043 |
- |
- |
- |
| 3 |
Hb_150651_020 |
0.1753657813 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_001001_120 |
0.1808393426 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_126933_020 |
0.1809433937 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 6 |
Hb_002043_100 |
0.1819869851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001907_080 |
0.1832205868 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 8 |
Hb_000186_110 |
0.2008337154 |
- |
- |
hypothetical protein CISIN_1g020639mg [Citrus sinensis] |
| 9 |
Hb_007894_190 |
0.2058430279 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 10 |
Hb_001597_020 |
0.2066096753 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 11 |
Hb_172426_060 |
0.20684922 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 12 |
Hb_004635_070 |
0.2089751372 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Solanum lycopersicum] |
| 13 |
Hb_024399_020 |
0.209955713 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 14 |
Hb_000062_480 |
0.2101077318 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005488_190 |
0.2161798724 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 16 |
Hb_001365_030 |
0.2169336181 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 17 |
Hb_000009_020 |
0.2171180122 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_004117_150 |
0.218476248 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 19 |
Hb_004807_050 |
0.2202178233 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 20 |
Hb_001942_020 |
0.2225170836 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |