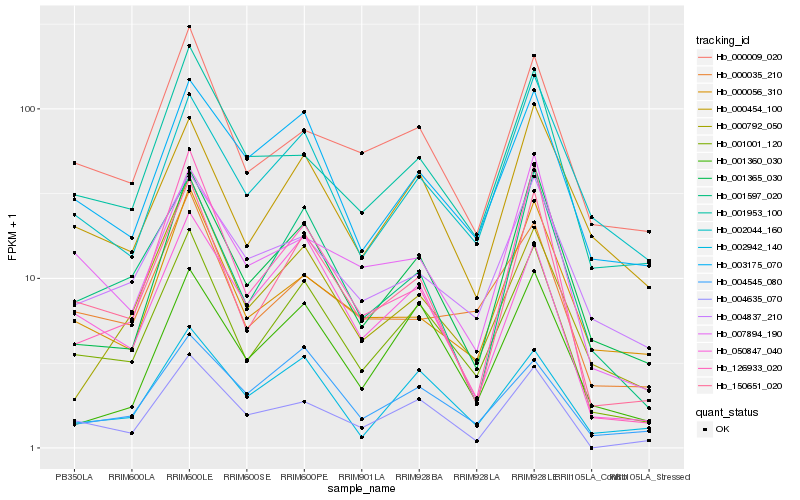
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001001_120 |
0.0 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 2 |
Hb_150651_020 |
0.0871861869 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_001365_030 |
0.1175093888 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 4 |
Hb_001597_020 |
0.1227143159 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004635_070 |
0.1245517821 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Solanum lycopersicum] |
| 6 |
Hb_002942_140 |
0.1261775786 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 7 |
Hb_003175_070 |
0.1263407748 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 8 |
Hb_000009_020 |
0.1270965847 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_050847_040 |
0.1274871591 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 10 |
Hb_000454_100 |
0.1289348254 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 11 |
Hb_001953_100 |
0.1292415443 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000035_210 |
0.1360157784 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 13 |
Hb_001360_030 |
0.1397937523 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 14 |
Hb_004837_210 |
0.1399220008 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 15 |
Hb_004545_080 |
0.1418173533 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000792_050 |
0.1443864201 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_002044_160 |
0.1449582738 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 18 |
Hb_007894_190 |
0.1451903873 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 19 |
Hb_000056_310 |
0.1462753233 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 20 |
Hb_126933_020 |
0.1470418902 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |