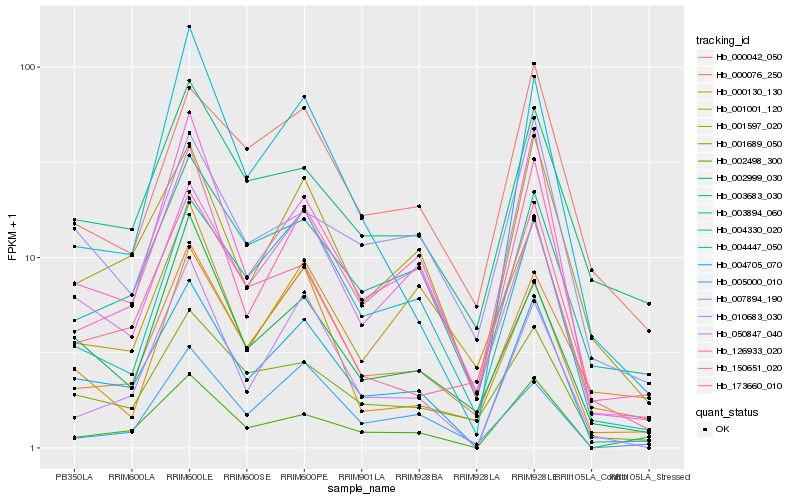
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004705_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647312 isoform X1 [Jatropha curcas] |
| 2 |
Hb_150651_020 |
0.108721878 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_001597_020 |
0.1206862113 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 4 |
Hb_050847_040 |
0.1388471331 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 5 |
Hb_126933_020 |
0.1448693715 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 6 |
Hb_010683_030 |
0.1451535333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003894_060 |
0.1466277544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004330_020 |
0.1466862863 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 9 |
Hb_002498_300 |
0.1467595676 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 10 |
Hb_001689_050 |
0.1507507624 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 11 |
Hb_002999_030 |
0.1557022676 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 12 |
Hb_003683_030 |
0.1583631479 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 13 |
Hb_004447_050 |
0.1603792278 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000042_050 |
0.1681405368 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001001_120 |
0.1683568778 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 16 |
Hb_007894_190 |
0.1683961811 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 17 |
Hb_000076_250 |
0.1687288789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000130_130 |
0.1699506574 |
- |
- |
hypothetical protein POPTR_0001s40330g [Populus trichocarpa] |
| 19 |
Hb_173660_010 |
0.1700628398 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Malus domestica] |
| 20 |
Hb_005000_010 |
0.1714607732 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |