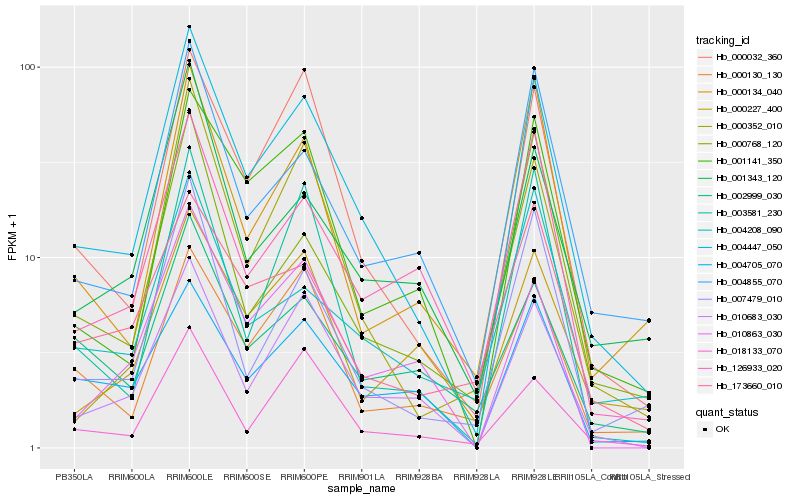
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004447_050 |
0.0 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000032_360 |
0.1137273326 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit S, chloroplastic [Jatropha curcas] |
| 3 |
Hb_010683_030 |
0.1215319936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001141_350 |
0.1351759703 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 5 |
Hb_007479_010 |
0.1441242203 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 6 |
Hb_004855_070 |
0.1441637243 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 7 |
Hb_000130_130 |
0.1522019818 |
- |
- |
hypothetical protein POPTR_0001s40330g [Populus trichocarpa] |
| 8 |
Hb_018133_070 |
0.1530535952 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 9 |
Hb_000352_010 |
0.1532372111 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000134_040 |
0.1535618061 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003581_230 |
0.153651644 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 12 |
Hb_126933_020 |
0.1553059019 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 13 |
Hb_010863_030 |
0.1592614624 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 14 |
Hb_004705_070 |
0.1603792278 |
- |
- |
PREDICTED: uncharacterized protein LOC105647312 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001343_120 |
0.1616460297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_173660_010 |
0.1631390707 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Malus domestica] |
| 17 |
Hb_002999_030 |
0.1668391609 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 18 |
Hb_004208_090 |
0.1672326507 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 19 |
Hb_000768_120 |
0.1673333711 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 20 |
Hb_000227_400 |
0.1675456559 |
- |
- |
lipoxygenase, putative [Ricinus communis] |