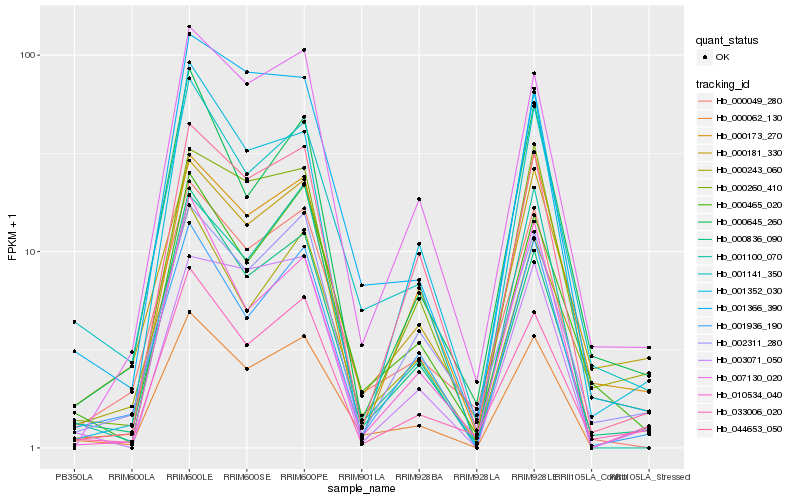
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_130 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_001141_350 |
0.1118497249 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 3 |
Hb_000836_090 |
0.1148123203 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 4 |
Hb_033006_020 |
0.1178167383 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 5 |
Hb_000181_330 |
0.1270224088 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 6 |
Hb_001936_190 |
0.1288827136 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 7 |
Hb_000243_060 |
0.1374153238 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 8 |
Hb_007130_020 |
0.1375350547 |
- |
- |
- |
| 9 |
Hb_002311_280 |
0.1407819473 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 10 |
Hb_010534_040 |
0.141308424 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003071_050 |
0.1424000365 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 12 |
Hb_000049_280 |
0.1426980244 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 13 |
Hb_000465_020 |
0.1454287518 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 14 |
Hb_000645_260 |
0.1461128647 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 15 |
Hb_044653_050 |
0.1463355503 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000260_410 |
0.1477516893 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 17 |
Hb_001366_390 |
0.1488316331 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 18 |
Hb_001352_030 |
0.1514883115 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 19 |
Hb_000173_270 |
0.1527090343 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001100_070 |
0.1532088218 |
- |
- |
PREDICTED: ferredoxin--nitrite reductase, chloroplastic isoform X1 [Jatropha curcas] |