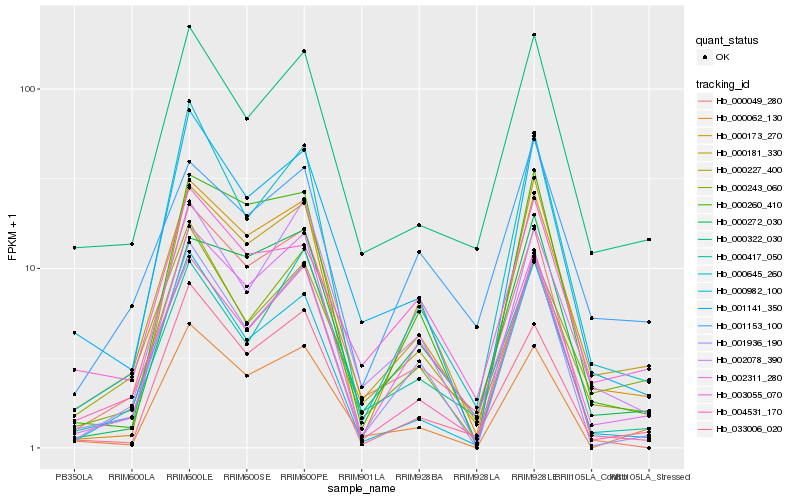
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_330 |
0.0 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 2 |
Hb_000243_060 |
0.091002341 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 3 |
Hb_000322_030 |
0.0914621783 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 4 |
Hb_002311_280 |
0.1014749821 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 5 |
Hb_000272_030 |
0.1130151456 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004531_170 |
0.1160054845 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 7 |
Hb_000173_270 |
0.116068664 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001141_350 |
0.1170605504 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 9 |
Hb_000227_400 |
0.1206821373 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 10 |
Hb_033006_020 |
0.1245498883 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 11 |
Hb_002078_390 |
0.1251377818 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 12 |
Hb_000260_410 |
0.1263258344 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 13 |
Hb_001153_100 |
0.1266102138 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 14 |
Hb_000062_130 |
0.1270224088 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_000645_260 |
0.1294569791 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 16 |
Hb_003055_070 |
0.1299966706 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_000417_050 |
0.1310785438 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 18 |
Hb_000049_280 |
0.1313543498 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 19 |
Hb_001936_190 |
0.1330288183 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 20 |
Hb_000982_100 |
0.1348097189 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PIF1-like isoform X1 [Jatropha curcas] |