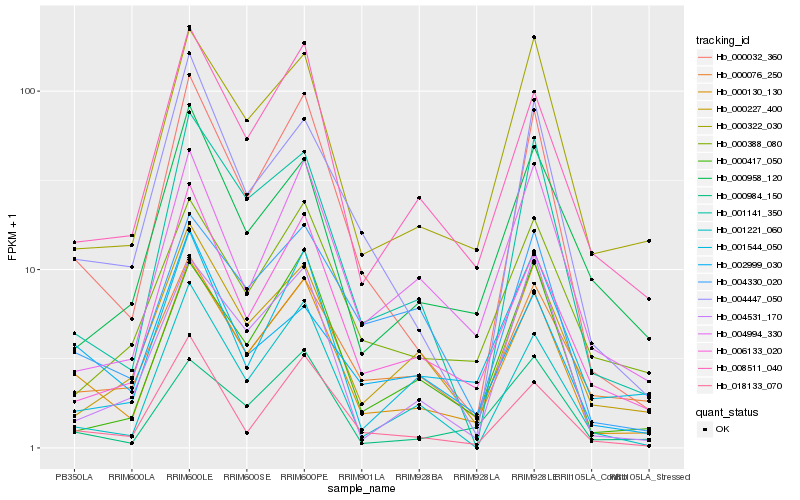
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_130 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s40330g [Populus trichocarpa] |
| 2 |
Hb_000032_360 |
0.1043623201 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit S, chloroplastic [Jatropha curcas] |
| 3 |
Hb_008511_040 |
0.1186846097 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_000322_030 |
0.1233032407 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 5 |
Hb_006133_020 |
0.1366434034 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 6 |
Hb_018133_070 |
0.138392937 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 7 |
Hb_000984_150 |
0.1400085005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001141_350 |
0.1407940089 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 9 |
Hb_000076_250 |
0.1409435223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000227_400 |
0.1486466926 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 11 |
Hb_001221_060 |
0.1494881366 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 12 |
Hb_004330_020 |
0.1511681631 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 13 |
Hb_004447_050 |
0.1522019818 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004994_330 |
0.1535699268 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000417_050 |
0.1537340724 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 16 |
Hb_000388_080 |
0.1567577271 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 17 |
Hb_001544_050 |
0.1584284351 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 18 |
Hb_002999_030 |
0.1598085085 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 19 |
Hb_004531_170 |
0.1607434095 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 20 |
Hb_000958_120 |
0.1614945563 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |