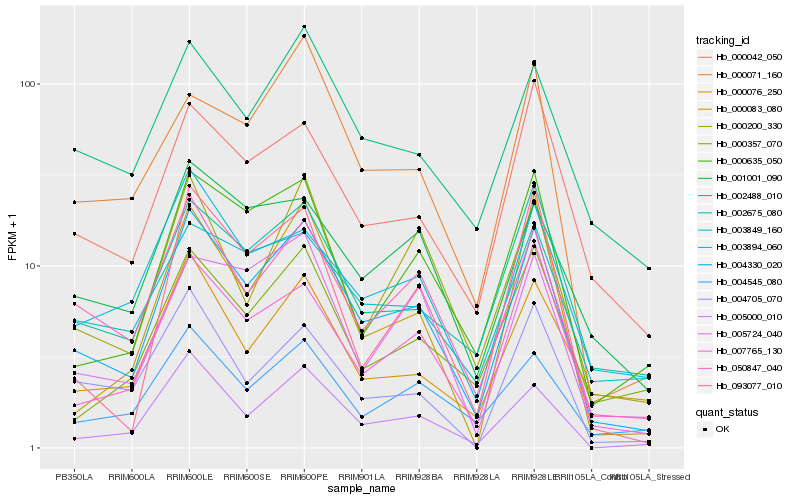
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004330_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 2 |
Hb_050847_040 |
0.1014887731 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 3 |
Hb_005000_010 |
0.1091592048 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002675_080 |
0.1262377468 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 5 |
Hb_007765_130 |
0.1264832263 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000635_050 |
0.1294465528 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 7 |
Hb_000042_050 |
0.132572872 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002488_010 |
0.1355212158 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 9 |
Hb_004545_080 |
0.1362575372 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_003894_060 |
0.1379466828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005724_040 |
0.1400169868 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 12 |
Hb_000083_080 |
0.1402052144 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 13 |
Hb_000076_250 |
0.1403000734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000200_330 |
0.1406106809 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 15 |
Hb_093077_010 |
0.1412668891 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001001_090 |
0.1430205309 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 17 |
Hb_003849_160 |
0.1442509452 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 18 |
Hb_000357_070 |
0.1448786483 |
- |
- |
- |
| 19 |
Hb_004705_070 |
0.1466862863 |
- |
- |
PREDICTED: uncharacterized protein LOC105647312 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000071_160 |
0.1467431738 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |