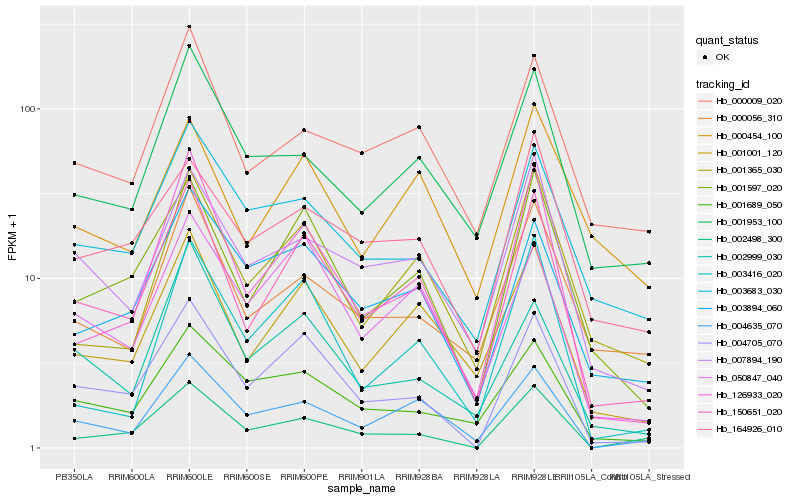
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150651_020 |
0.0 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 2 |
Hb_001001_120 |
0.0871861869 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 3 |
Hb_001597_020 |
0.09128582 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004705_070 |
0.108721878 |
- |
- |
PREDICTED: uncharacterized protein LOC105647312 isoform X1 [Jatropha curcas] |
| 5 |
Hb_126933_020 |
0.1145830739 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 6 |
Hb_004635_070 |
0.1232876816 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Solanum lycopersicum] |
| 7 |
Hb_007894_190 |
0.1286766262 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 8 |
Hb_001365_030 |
0.1307881338 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 9 |
Hb_050847_040 |
0.1311726887 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 10 |
Hb_000009_020 |
0.1354809519 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_003894_060 |
0.1464208042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000056_310 |
0.1467043944 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 13 |
Hb_001953_100 |
0.147830174 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000454_100 |
0.1507306793 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 15 |
Hb_164926_010 |
0.1538573357 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 16 |
Hb_003683_030 |
0.1547519054 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 17 |
Hb_003416_020 |
0.1551449986 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 18 |
Hb_002999_030 |
0.1555532937 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 19 |
Hb_002498_300 |
0.1555755369 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 20 |
Hb_001689_050 |
0.1566810087 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |