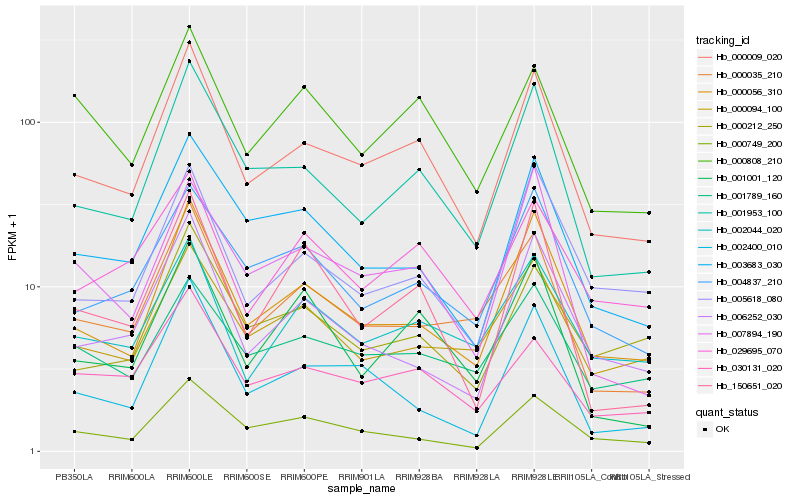
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_020 |
0.0 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001953_100 |
0.0836504158 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000056_310 |
0.0870298468 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 4 |
Hb_030131_020 |
0.1128434468 |
- |
- |
Carboxypeptidase B2 precursor, putative [Ricinus communis] |
| 5 |
Hb_000035_210 |
0.1146878108 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 6 |
Hb_003683_030 |
0.1169922183 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 7 |
Hb_004837_210 |
0.1194302234 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 8 |
Hb_002044_020 |
0.1238435017 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 9 |
Hb_005618_080 |
0.1244236641 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000749_200 |
0.1250839268 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001001_120 |
0.1270965847 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_007894_190 |
0.1276244459 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 13 |
Hb_029695_070 |
0.1276981178 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_006252_030 |
0.127968964 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 15 |
Hb_002400_010 |
0.1306090355 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 16 |
Hb_000212_250 |
0.1310217592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000808_210 |
0.1326082088 |
- |
- |
hypothetical protein EUTSA_v10023481mg [Eutrema salsugineum] |
| 18 |
Hb_000094_100 |
0.1332279651 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 19 |
Hb_001789_160 |
0.1336393028 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 20 |
Hb_150651_020 |
0.1354809519 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |