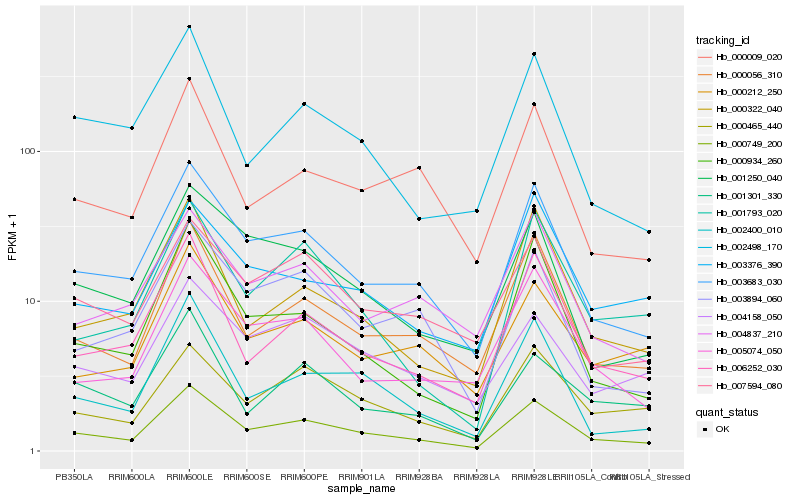
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_200 |
0.0 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003683_030 |
0.0694436731 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 3 |
Hb_000056_310 |
0.0987094203 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 4 |
Hb_006252_030 |
0.1036214583 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 5 |
Hb_000465_440 |
0.1149213299 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 6 |
Hb_002400_010 |
0.1199520841 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 7 |
Hb_000934_260 |
0.1203687502 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_005074_050 |
0.1218379695 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 9 |
Hb_007594_080 |
0.1223487722 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 10 |
Hb_001250_040 |
0.1232366491 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 11 |
Hb_000322_040 |
0.1237889905 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004158_050 |
0.1242170373 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 13 |
Hb_000212_250 |
0.1248478682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000009_020 |
0.1250839268 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_003894_060 |
0.1274852651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002498_170 |
0.1276442289 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_004837_210 |
0.1288033878 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 18 |
Hb_001301_330 |
0.1289931462 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 19 |
Hb_001793_020 |
0.1304797523 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 20 |
Hb_003376_390 |
0.1314971292 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |