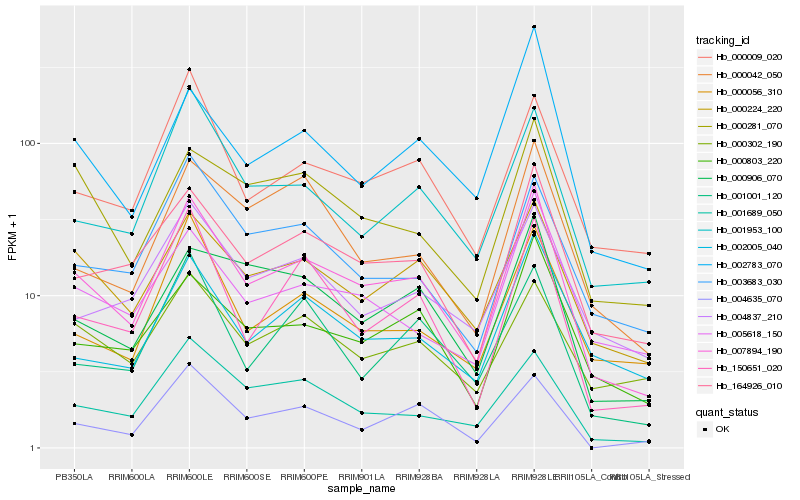
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 2 |
Hb_164926_010 |
0.1030333811 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 3 |
Hb_000224_220 |
0.1172209131 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |
| 4 |
Hb_000803_220 |
0.119544768 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001689_050 |
0.1249934158 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 6 |
Hb_000009_020 |
0.1276244459 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_150651_020 |
0.1286766262 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_001953_100 |
0.1291666897 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005618_150 |
0.1298731289 |
- |
- |
PREDICTED: protein translocase subunit SecA, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000056_310 |
0.1318137318 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 11 |
Hb_002005_040 |
0.1338934606 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_003683_030 |
0.1365459231 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 13 |
Hb_000906_070 |
0.1373653072 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_004635_070 |
0.1373840751 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Solanum lycopersicum] |
| 15 |
Hb_002783_070 |
0.137540845 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 16 |
Hb_004837_210 |
0.1386112294 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 17 |
Hb_000281_070 |
0.141077701 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 18 |
Hb_000042_050 |
0.1436469666 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000302_190 |
0.1443363966 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 20 |
Hb_001001_120 |
0.1451903873 |
- |
- |
protein phosphatase, putative [Ricinus communis] |