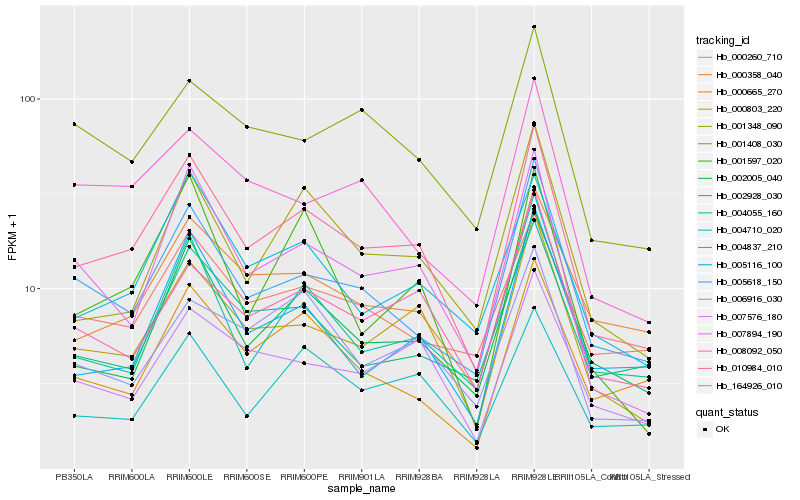
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164926_010 |
0.0 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 2 |
Hb_000803_220 |
0.0872288417 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007894_190 |
0.1030333811 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 4 |
Hb_002005_040 |
0.1082954055 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_005618_150 |
0.1092296431 |
- |
- |
PREDICTED: protein translocase subunit SecA, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004837_210 |
0.1096909609 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 7 |
Hb_007576_180 |
0.1165545744 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 8 |
Hb_000358_040 |
0.117342644 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_010984_010 |
0.1179320393 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001348_090 |
0.1219658585 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004710_020 |
0.1231553625 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 12 |
Hb_005116_100 |
0.1248999061 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 13 |
Hb_000260_710 |
0.1259739095 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006916_030 |
0.1279205438 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 15 |
Hb_004055_160 |
0.12898551 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002928_030 |
0.1292690811 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 17 |
Hb_008092_050 |
0.1293949217 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 18 |
Hb_000665_270 |
0.1297708041 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 19 |
Hb_001597_020 |
0.130802628 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001408_030 |
0.1313272561 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |