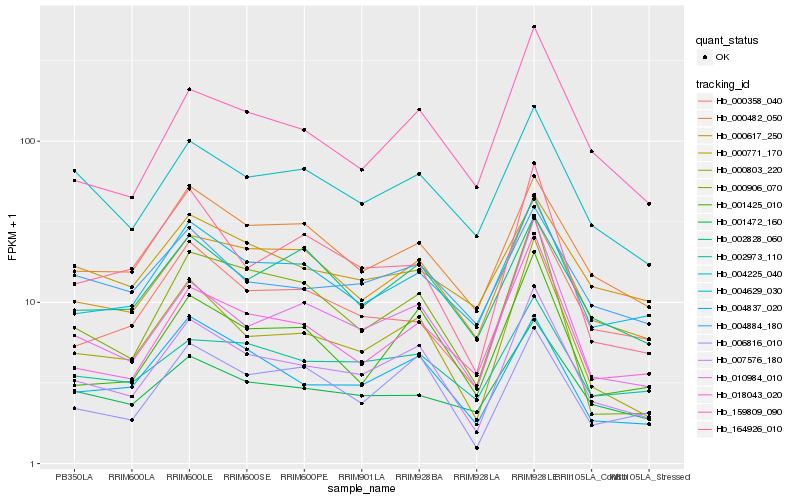
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007576_180 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 2 |
Hb_000803_220 |
0.0676437033 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 3 |
Hb_010984_010 |
0.0892716948 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_004629_030 |
0.1001273125 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_004837_020 |
0.1015618218 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |
| 6 |
Hb_001425_010 |
0.1021171489 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_018043_020 |
0.10385253 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006816_010 |
0.1043905376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_159809_090 |
0.1075200944 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 10 |
Hb_004225_040 |
0.1095849761 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 11 |
Hb_000482_050 |
0.1104013132 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000358_040 |
0.114368628 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_164926_010 |
0.1165545744 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 14 |
Hb_002973_110 |
0.1189009588 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_004884_180 |
0.1199004756 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001472_160 |
0.1211138401 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 17 |
Hb_002828_060 |
0.1216935692 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000771_170 |
0.1221461176 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_000617_250 |
0.1223692621 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000906_070 |
0.1226064641 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |