| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
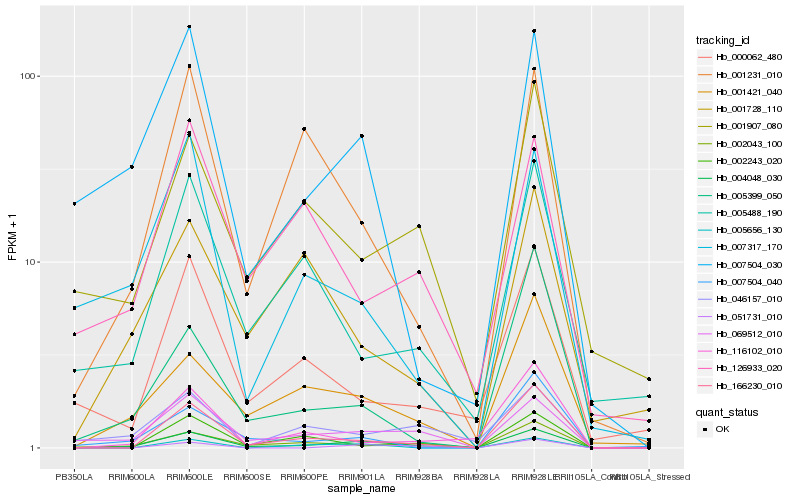
Hb_004048_030 |
0.0 |
- |
- |
hypothetical protein B456_009G371200, partial [Gossypium raimondii] |
| 2 |
Hb_069512_010 |
0.1835888496 |
- |
- |
- |
| 3 |
Hb_002043_100 |
0.1847700522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001421_040 |
0.1948365746 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 2 [Jatropha curcas] |
| 5 |
Hb_166230_010 |
0.2032712224 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_007504_030 |
0.2156145587 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 7 |
Hb_001231_010 |
0.2283404449 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 8 |
Hb_005656_130 |
0.2410906079 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 9 |
Hb_001907_080 |
0.245000192 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 10 |
Hb_007504_040 |
0.2467721988 |
- |
- |
hypothetical chloroplast RF2 [Fragaria iinumae] |
| 11 |
Hb_046157_010 |
0.2469062912 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 12 |
Hb_051731_010 |
0.2470841877 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 13 |
Hb_007317_170 |
0.2478453147 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 14 |
Hb_002243_020 |
0.2487017451 |
- |
- |
PREDICTED: uncharacterized protein LOC105642097 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000062_480 |
0.2495334286 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 16 |
Hb_126933_020 |
0.2521302189 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 17 |
Hb_005488_190 |
0.253852424 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 18 |
Hb_001728_110 |
0.2546712876 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF039-like [Jatropha curcas] |
| 19 |
Hb_116102_010 |
0.2560428247 |
- |
- |
hypothetical protein POPTR_0204s00240g [Populus trichocarpa] |
| 20 |
Hb_005399_050 |
0.2571744778 |
- |
- |
DNA-directed RNA polymerase subunit beta [Glycine soja] |