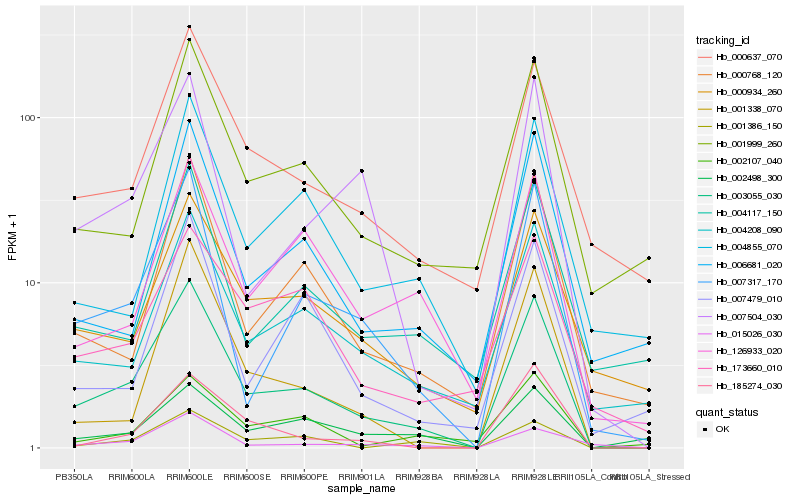
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003055_030 |
0.0 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 2 |
Hb_007317_170 |
0.1353936656 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 3 |
Hb_004208_090 |
0.1637143441 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 4 |
Hb_000768_120 |
0.1640105628 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 5 |
Hb_000637_070 |
0.1658842625 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 6 |
Hb_015026_030 |
0.1742911913 |
- |
- |
hypothetical protein POPTR_0011s04350g [Populus trichocarpa] |
| 7 |
Hb_001338_070 |
0.174612087 |
- |
- |
ORF126 [Jatropha curcas] |
| 8 |
Hb_001999_260 |
0.1754834465 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_007504_030 |
0.1756735178 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 10 |
Hb_000934_260 |
0.1767445385 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_006681_020 |
0.1782977755 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001386_150 |
0.1787372937 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_126933_020 |
0.1827458888 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 14 |
Hb_185274_030 |
0.187629791 |
- |
- |
hypothetical protein JCGZ_15170 [Jatropha curcas] |
| 15 |
Hb_004117_150 |
0.1892455944 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 16 |
Hb_004855_070 |
0.1893939571 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 17 |
Hb_002107_040 |
0.19177771 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 18 |
Hb_007479_010 |
0.1921460958 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 19 |
Hb_002498_300 |
0.1943914882 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 20 |
Hb_173660_010 |
0.1948704237 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Malus domestica] |