| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
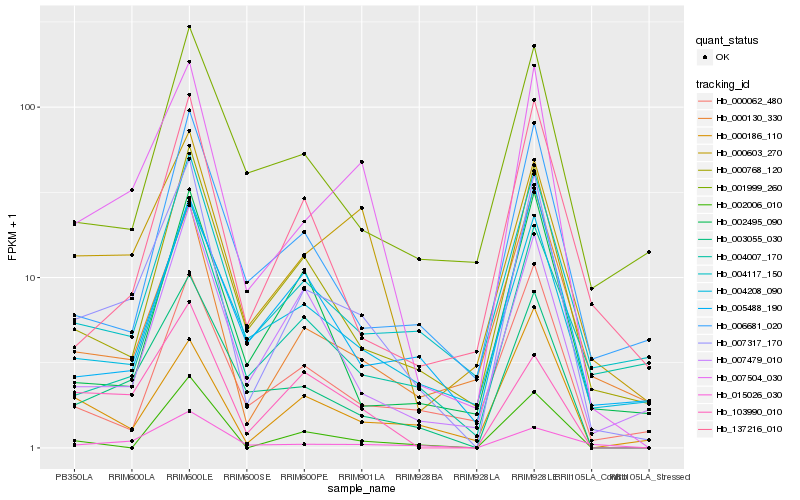
Hb_007317_170 |
0.0 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 2 |
Hb_007504_030 |
0.1211563087 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 3 |
Hb_003055_030 |
0.1353936656 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 4 |
Hb_000768_120 |
0.1550358112 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 5 |
Hb_004117_150 |
0.1735424206 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 6 |
Hb_004208_090 |
0.1751067483 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 7 |
Hb_000130_330 |
0.1755364149 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |
| 8 |
Hb_007479_010 |
0.1780773089 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 9 |
Hb_015026_030 |
0.1836128843 |
- |
- |
hypothetical protein POPTR_0011s04350g [Populus trichocarpa] |
| 10 |
Hb_006681_020 |
0.1853877799 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 11 |
Hb_137216_010 |
0.1893608043 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 12 |
Hb_000062_480 |
0.1925414051 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000603_270 |
0.1933489371 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 14 |
Hb_002006_010 |
0.194424616 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 15 |
Hb_005488_190 |
0.1982553112 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 16 |
Hb_002495_090 |
0.1991211735 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004007_170 |
0.200547894 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 18 |
Hb_001999_260 |
0.2013822813 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_103990_010 |
0.2018043895 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 20 |
Hb_000186_110 |
0.2018737857 |
- |
- |
hypothetical protein CISIN_1g020639mg [Citrus sinensis] |