| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
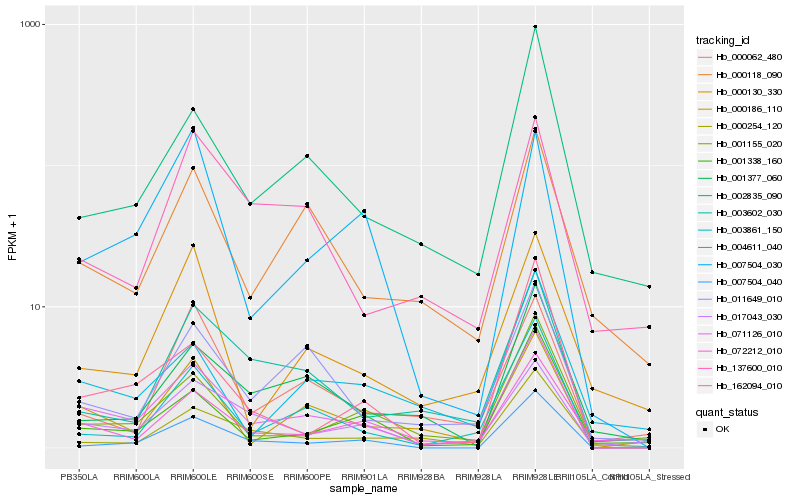
| 1 |
Hb_071126_010 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_017043_030 |
0.1637544623 |
- |
- |
hypothetical protein CISIN_1g011604mg [Citrus sinensis] |
| 3 |
Hb_000186_110 |
0.1661681034 |
- |
- |
hypothetical protein CISIN_1g020639mg [Citrus sinensis] |
| 4 |
Hb_003861_150 |
0.1679386849 |
- |
- |
hypothetical protein JCGZ_15167 [Jatropha curcas] |
| 5 |
Hb_011649_010 |
0.1809425999 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 6 |
Hb_072212_010 |
0.1814543164 |
- |
- |
hypothetical chloroplast RF1 [Ficus sp. M. J. Moore 315] |
| 7 |
Hb_000118_090 |
0.1866482246 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_002835_090 |
0.1870778193 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 9 |
Hb_003602_030 |
0.1927719182 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 10 |
Hb_007504_040 |
0.1958470811 |
- |
- |
hypothetical chloroplast RF2 [Fragaria iinumae] |
| 11 |
Hb_001377_060 |
0.2026140765 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 12 |
Hb_007504_030 |
0.2044603846 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 13 |
Hb_001338_160 |
0.2048508578 |
- |
- |
NADH dehydrogenase subunit 5 [Hevea brasiliensis] |
| 14 |
Hb_001155_020 |
0.2050089463 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_162094_010 |
0.2068287421 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 16 |
Hb_000062_480 |
0.2074253667 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 17 |
Hb_137600_010 |
0.2092984378 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 18 |
Hb_004611_040 |
0.2106002245 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigD, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000254_120 |
0.211488653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000130_330 |
0.211951968 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |