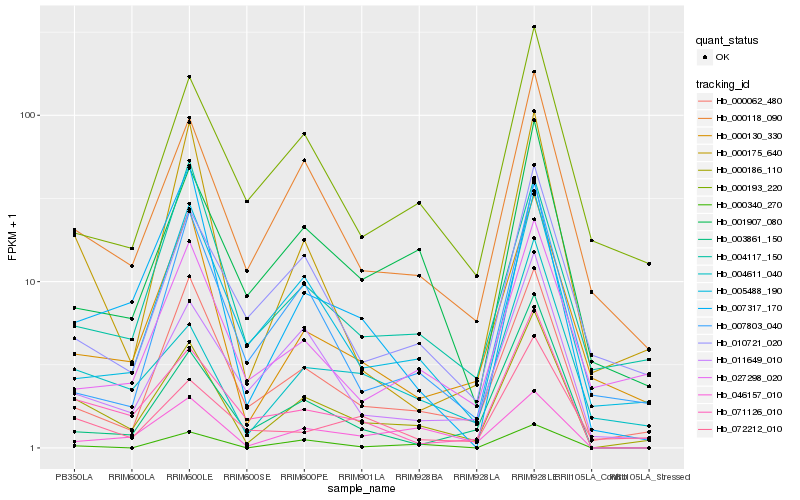
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_110 |
0.0 |
- |
- |
hypothetical protein CISIN_1g020639mg [Citrus sinensis] |
| 2 |
Hb_000118_090 |
0.152892987 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000062_480 |
0.1630347538 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 4 |
Hb_071126_010 |
0.1661681034 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000130_330 |
0.1665055115 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |
| 6 |
Hb_000175_640 |
0.1682906951 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004611_040 |
0.1757513232 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigD, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_011649_010 |
0.1772799731 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 9 |
Hb_001907_080 |
0.1809234295 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 10 |
Hb_005488_190 |
0.1877934477 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 11 |
Hb_000193_220 |
0.19304275 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_010721_020 |
0.1953658526 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_007803_040 |
0.1960364159 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 14 |
Hb_072212_010 |
0.1977451101 |
- |
- |
hypothetical chloroplast RF1 [Ficus sp. M. J. Moore 315] |
| 15 |
Hb_046157_010 |
0.2008337154 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 16 |
Hb_007317_170 |
0.2018737857 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 17 |
Hb_000340_270 |
0.2019834524 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 18 |
Hb_027298_020 |
0.2021130523 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 19 |
Hb_003861_150 |
0.202456225 |
- |
- |
hypothetical protein JCGZ_15167 [Jatropha curcas] |
| 20 |
Hb_004117_150 |
0.202776924 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |