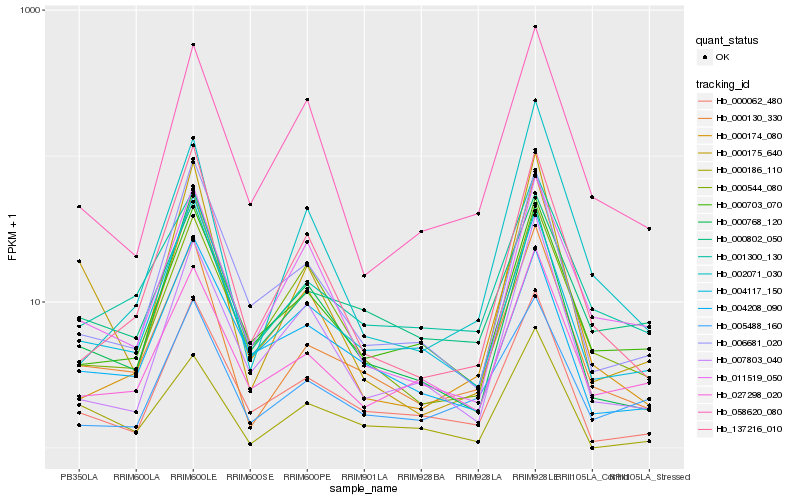
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_330 |
0.0 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |
| 2 |
Hb_001300_130 |
0.1207471422 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 3 |
Hb_004117_150 |
0.1326902899 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 4 |
Hb_000062_480 |
0.1360727141 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 5 |
Hb_027298_020 |
0.1376837798 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 6 |
Hb_000703_070 |
0.1407023428 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000802_050 |
0.1421518361 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 8 |
Hb_137216_010 |
0.1431972979 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 9 |
Hb_005488_160 |
0.1464038304 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000174_080 |
0.1480045197 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 11 |
Hb_002071_030 |
0.1534062914 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |
| 12 |
Hb_000175_640 |
0.1542052708 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_006681_020 |
0.1578188335 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000768_120 |
0.160993013 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 15 |
Hb_000544_080 |
0.1662402849 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 16 |
Hb_004208_090 |
0.1662404394 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 17 |
Hb_000186_110 |
0.1665055115 |
- |
- |
hypothetical protein CISIN_1g020639mg [Citrus sinensis] |
| 18 |
Hb_011519_050 |
0.1669610396 |
- |
- |
hypothetical protein POPTR_0015s04810g [Populus trichocarpa] |
| 19 |
Hb_058620_080 |
0.1677084953 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007803_040 |
0.1682469736 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |