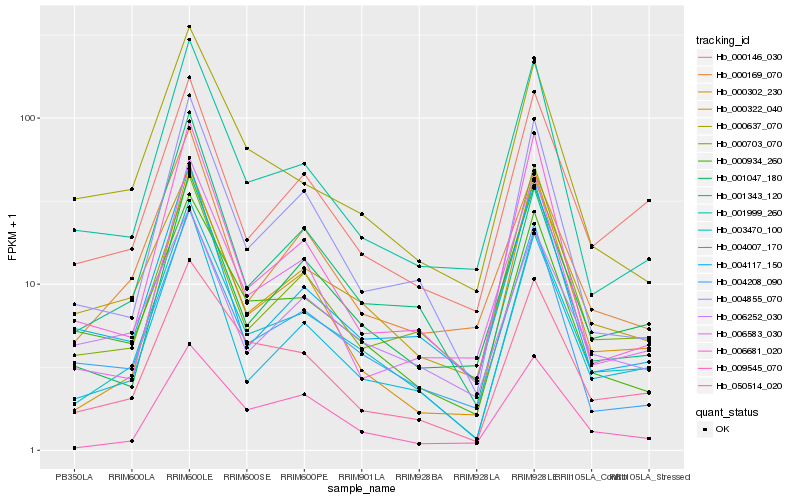
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003470_100 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_004007_170 |
0.0795835176 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_050514_020 |
0.0987201282 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 4 |
Hb_004855_070 |
0.1104118714 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 5 |
Hb_001999_260 |
0.1125168543 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004208_090 |
0.1151167932 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 7 |
Hb_001047_180 |
0.116398648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000934_260 |
0.1177770045 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000146_030 |
0.1182819509 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000169_070 |
0.1207039405 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 11 |
Hb_000322_040 |
0.1219202718 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006681_020 |
0.1263155546 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 13 |
Hb_009545_070 |
0.1274382987 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Jatropha curcas] |
| 14 |
Hb_000302_230 |
0.1278422024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000637_070 |
0.1281278461 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 16 |
Hb_004117_150 |
0.129346862 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 17 |
Hb_006252_030 |
0.1327491308 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 18 |
Hb_006583_030 |
0.1336312162 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 19 |
Hb_000703_070 |
0.140251243 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001343_120 |
0.1409257022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |