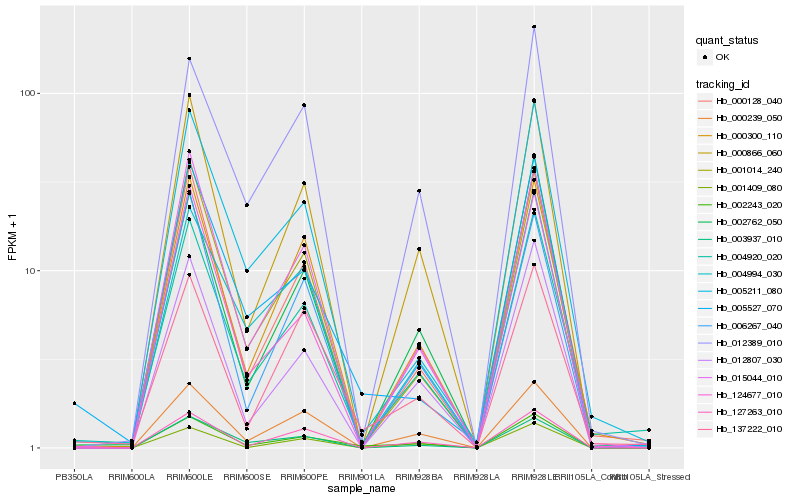
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002243_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642097 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000866_060 |
0.1101043139 |
- |
- |
putative glycerol-3-phosphate acyltransferase [Vernicia fordii] |
| 3 |
Hb_004920_020 |
0.1170563027 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 4 |
Hb_000239_050 |
0.1207438836 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |
| 5 |
Hb_127263_010 |
0.1232708391 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 6 |
Hb_012807_030 |
0.1236458653 |
- |
- |
PREDICTED: uncharacterized protein LOC105628042 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000300_110 |
0.1245161094 |
- |
- |
hypothetical protein JCGZ_02651 [Jatropha curcas] |
| 8 |
Hb_000128_040 |
0.1248778834 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 9 |
Hb_015044_010 |
0.1250917575 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 10 |
Hb_124677_010 |
0.1273435739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004994_030 |
0.127970098 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 12 |
Hb_001014_240 |
0.1280737771 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 13 |
Hb_137222_010 |
0.1298059686 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 14 |
Hb_003937_010 |
0.1300396246 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 15 |
Hb_002762_050 |
0.1329246581 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 16 |
Hb_012389_010 |
0.1330904364 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 17 |
Hb_001409_080 |
0.1338580133 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Populus euphratica] |
| 18 |
Hb_005527_070 |
0.135216261 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 19 |
Hb_006267_040 |
0.1356955659 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005211_080 |
0.1371790858 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |