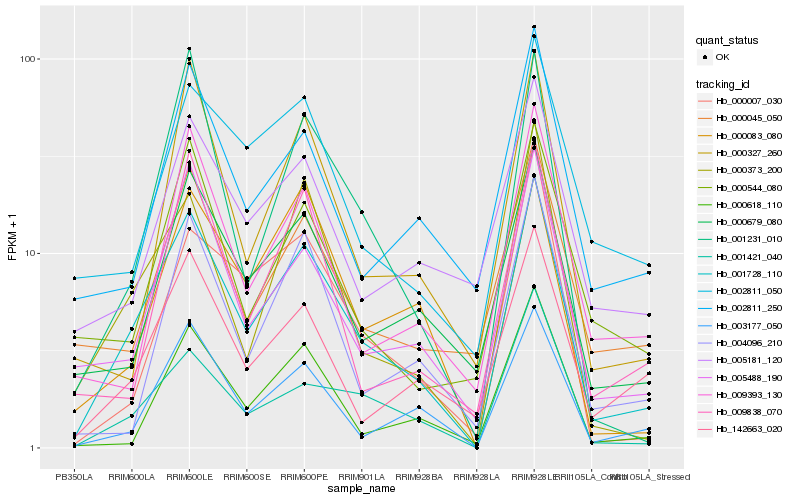
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001728_110 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF039-like [Jatropha curcas] |
| 2 |
Hb_001421_040 |
0.1438820225 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 2 [Jatropha curcas] |
| 3 |
Hb_003177_050 |
0.147451678 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Populus euphratica] |
| 4 |
Hb_005488_190 |
0.149625763 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 5 |
Hb_000373_200 |
0.1506596416 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 6 |
Hb_000679_080 |
0.1543726574 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001231_010 |
0.1568305175 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 8 |
Hb_000618_110 |
0.1639312035 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 9 |
Hb_142663_020 |
0.1642079965 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 10 |
Hb_000327_260 |
0.1716499771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000045_050 |
0.1719848547 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_009393_130 |
0.1720263068 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 13 |
Hb_004096_210 |
0.1723109896 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002811_250 |
0.1728855148 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000083_080 |
0.1740773309 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 16 |
Hb_002811_050 |
0.1747298167 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 17 |
Hb_009838_070 |
0.1760071 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000544_080 |
0.176099102 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000007_030 |
0.1762394435 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_005181_120 |
0.1769776723 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |